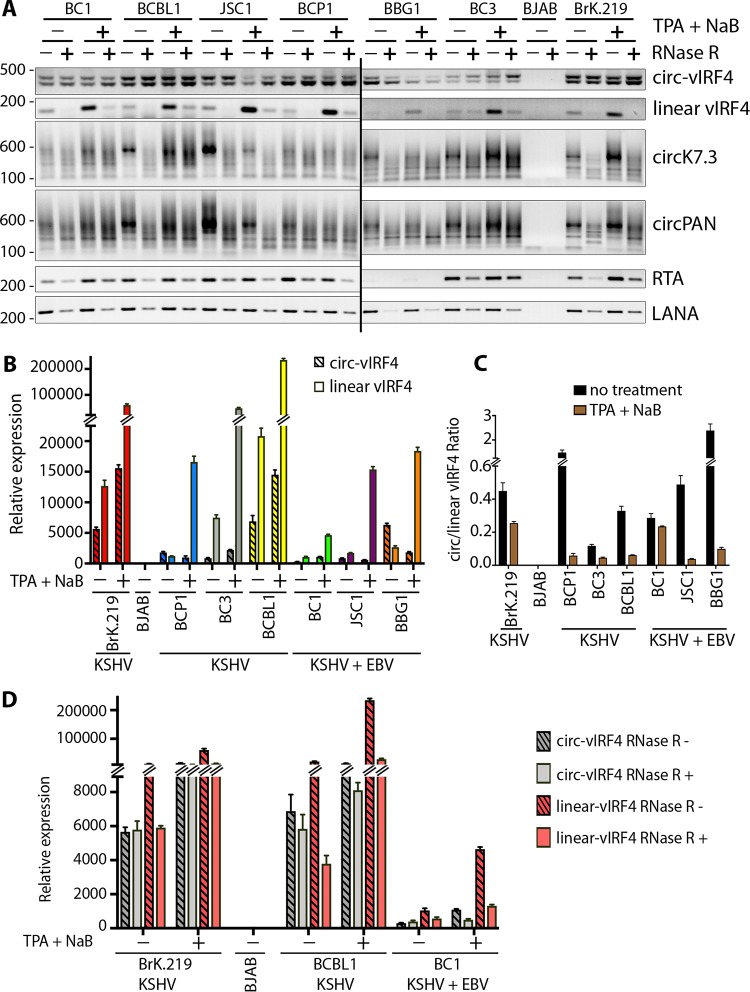
FIG 1.
Expression profile of KSHV circRNAs in a panel of PEL cells. Total RNA was extracted from PEL cells with or without induction of the KSHV lytic cycle using a combination of TPA and NaB. (A) RT-PCR was performed on RNase R-treated (+) or untreated (-) samples using divergent primer pairs corresponding to circ-vIRF4, circPANs, or circK7.3, while linear transcripts (vIRF4, LANA, and RTA) were amplified using a pair of convergent primers specific to each gene. (B) Quantitative comparison of circular and linear vIRF4 expression in cDNA from RNase R (-) samples processed as described for panel A using a TaqMan-based qPCR with a probe spanning circ-vIRF4 junction and a probe corresponding to the 3′ end of linear vIRF4, outside the circRNA coding sequence. (C) Ratio of circ-vIRF4 expression to linear vIRF4 expression in the experiment described for panel B. (D) Comparison of circ-vIRF4 expression and linear vIRF4 expression in the presence or absence of RNase R treatment. CT values were normalized with the RNase P CT value to calculate delta CT, and all samples were normalized to BJAB cells to calculate delta-delta CT (relative expression) values. Bar graphs in panels B, C, and D represent means ± standard deviations (SD) of results from three replicates.