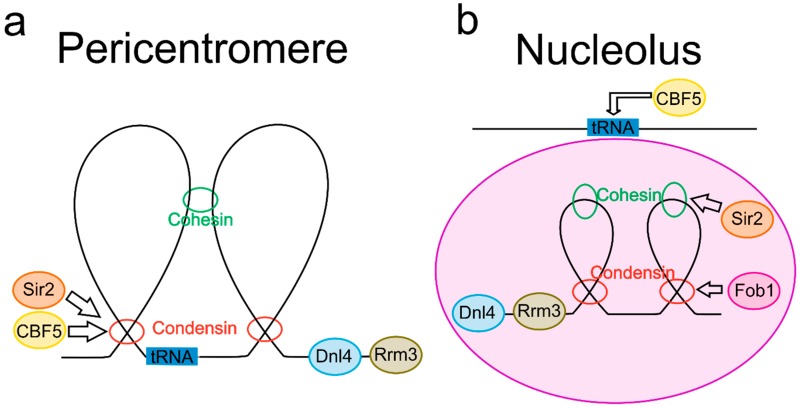
Figure 1.
DNA loops in the pericentromere and nucleolus. (a) Pericentromere loop schematic. Condensin extrudes DNA in the pericentromere [12,17,18,34] while cohesin radially links nearby loops [11,12]. Pericentric condensin enrichment is controlled by both Cbf5, a small nucleolar ribonucleoprotein, [35] and the histone deacetylase Sir2 [17]. DNA helicase Rrm3 regulates replication fork stalling at the pericentromere [36], and DNA ligase 4 (Dnl4) regulates segregation with a potential role in pericentric fork stalling as well [37,38]. tRNA genes are located in the pericentromere [39,40,41], and are associated with both condensin and cohesin [5,41,42]. (b) Nucleolus loop schematic. Condensin [43,44,45,46] and cohesin [23] both regulate loop formation in rDNA. DNA replication fork blocking protein (Fob1) regulates enrichment of condensin in rDNA [47], whereas Sir2 regulates cohesin rDNA localization [48]. Dnl4 [37] and Rrm3 [36] both control fork stalling at rDNA repeats. tRNA genes are tethered to the nucleolus in a Cbf5-dependent manner [49].