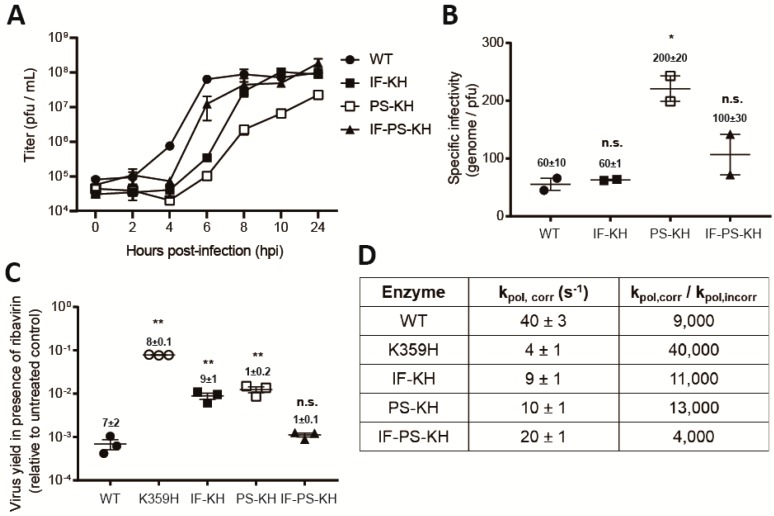
Figure 2.
Biological and biochemical characterization of variants of K359H PV and the RdRp reveal that both substitutions are required to achieve a near wild-type phenotype. (A) One-step growth analysis. Cells were infected at MOI 10 with the following PVs: WT, IF-KH, PS-KH, and IF-PS-KH. Viral titer (pfu/mL) was plotted as a function of time post-infection. Duplicate infected samples were used for plaque assays. Error bars indicate SEM (n = 2), biological replicates. (B) Specific infectivity. Virus was isolated 24 h post-infection and used for qRT-PCR to determine genomes/mL or plaque assay to determine pfu/mL, with the quotient yielding specific infectivity, genomes/pfu. Two biological replicates performed in triplicate for qPCR analysis. Statistical analyses were performed using unpaired, two-tailed t-test (* significance level p ≤ 0.05, n.s. = non-significant). (C) Ribavirin sensitivity. HeLa cells were infected at a MOI 0.1 with each PV in the presence of 600 µM ribavirin. After a 24-h incubation at 37 °C, virus was isolated and used for plaque assay. Indicated is the titer of virus recovered in the presence of ribavirin normalized to that recovered in the absence of ribavirin. Solid bar indicates the mean of each virus yield. Error bars indicate SEM (n = 3). Statistical analyses were performed using unpaired, two-tailed t-test (** significance level p ≤ 0.005, n.s. = non-significant). (D) Biochemical analysis. The rate constant for incorporation of a single correct nucleotide, ATP, (kpol,corr) and a single incorrect nucleotide, GTP, (kpol,incorr) by each PV RdRp was performed as previously described [18]. Data are reported using one significant figure. The error reported is the standard error from the fit of the data to a single exponential [18].