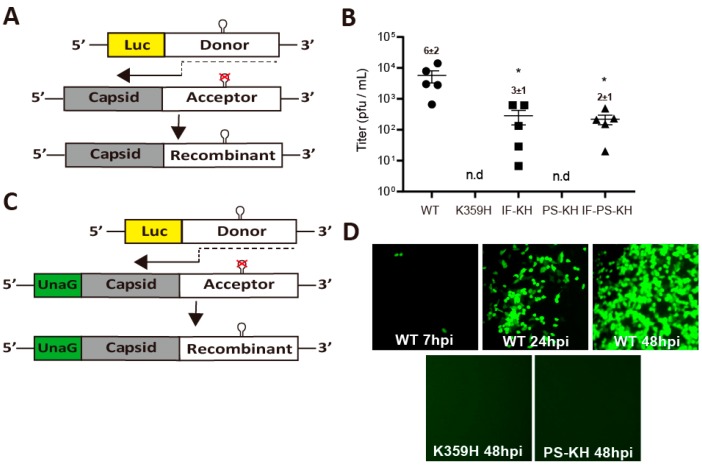
Figure 3.
Parameters in addition to RdRp speed and fidelity contribute to recombination efficiency. (A) Schematic of the PV recombination assay used [31]. Two RNAs are used: a replication-competent subgenomic RNA lacking capsid-coding sequence (donor RNA); and a replication-incompetent full-length genomic RNA with a defective cis-acting replication element (CRE, indicated by hairpin with defective version indicated by X) in 2C-coding sequence (acceptor RNA). Co-transfection of these RNAs produces infectious virus if recombination occurs. (B) The indicated PV RdRp was engineered into both donor and accepter RNA and co-transfected into a L929 mouse fibroblast cell line. Infectious virus produced by recombination in L929 cells was determined by plaque assay using HeLa cells. Each point shown is an independent experiment reflecting the average of three replicates. When plaques could not be detected, n.d. is indicated. Mean and SEM (n = 5) are indicated. Statistical analyses were performed using unpaired, two-tailed t-test (* significance level p ≤ 0.05). (C) Schematic of a modified PV recombination assay using an acceptor RNA expressing the green fluorescent protein, UnaG [28]. Recombinants are scored by expression of green fluorescence instead of plaques, thereby increasing the sensitivity. (D) Infectious virus produced by recombination in L929 was scored for infectious virus in HeLa cells 7, 24, and 48 h post-infection for WT and at 48 h only for K359H and PS-KH. Shown is one representative image from three independent experiments.