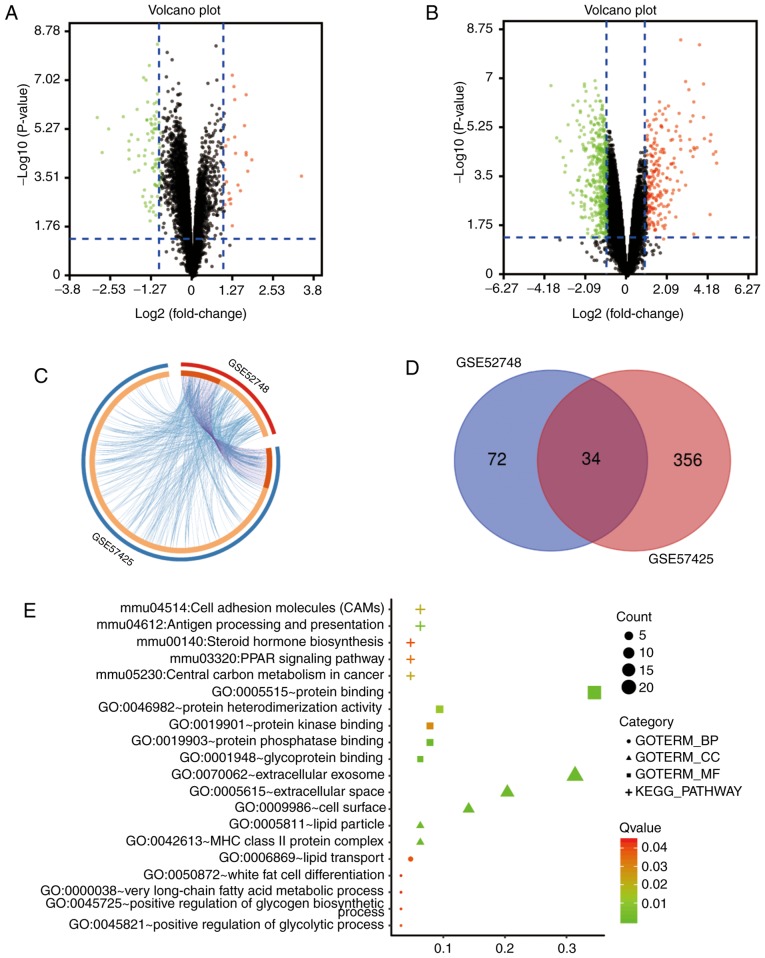
Figure 1.
Bioinformatics analysis of NAFLD-related datasets. (A and B) DEGs identified from the GEO database from the datasets (A) GSE52748 and (B) GSE57425. Upregulated genes are indicated as red dots based on log fold-change >2.0 and P<0.05. Downregulated genes are indicated as green dots based on log fold-change <2.0 and P<0.05. Genes without significant differences are indicated as black dots. (C) Overlap analysis between gene lists and shared enriched ontologies. The Circos plot depicts the overlap between genes and functional categories based on two input gene lists derived from the DEGs. The outer circle represents the identity of the corresponding gene list, labeled GSE52748 and GSE57425, and the inner circle represents gene lists, labeled the 462 DEGs in the two data sets, where hits are arranged along the arc. Genes on the two lists are indicated in dark orange and genes unique to a list are presented as light orange. Purple curves link identical genes and blue curves link genes that belong to the same enriched ontology term. (D) Venn diagram of DEGs in the two GEO datasets. (E) GO and KEGG pathway enrichment analysis of overlapping DEGs in the two GEO datasets. GO analysis included three functional groups: Molecular functions, biological processes and cell components. DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; NAFLD, non-alcoholic fatty liver disease.