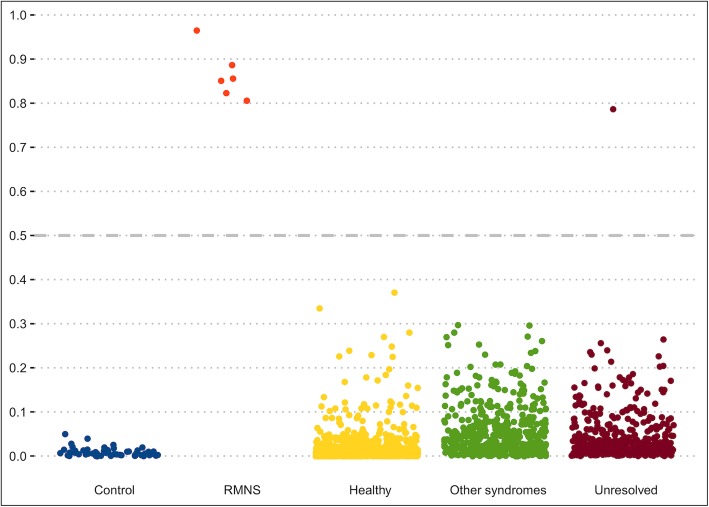
Fig. 2.
A classification model using DNA methylation data yields full sensitivity and specificity in classifying patients with Rahman syndrome. Each panel on the x-axis illustrates testing for a group of subjects with a distinct phenotype, as indicated on bottom of the panel. Y-axis represents scores generated by the classifier for different subjects as indicated by points on the plot. The scores range 0–1, with higher scores indicating a higher chance of having a methylation profile similar to Rahman syndrome (RMNS) (y-axis). By default, the classifier utilizes a cutoff of 0.5 for assigning the class; however, the vast majority of the tested individuals received a score close to 0 or 1. Therefore, for the purpose of better visualization, the points are jittered. Control (blue): 60 controls used to describe the signature and train the model; RMNS (red): six patients with RMNS used for identification of the episignature and training of the classifier; Healthy (yellow): 1678 controls used to measure the specificity of the model; Other syndromes (green): 502 patients with confirmed clinical and molecular diagnosis of various Mendelian disorders resulting from defects in epigenetic machinery; Unresolved (maroon): 453 patients with developmental abnormalities but without a diagnosis at the time of assessment