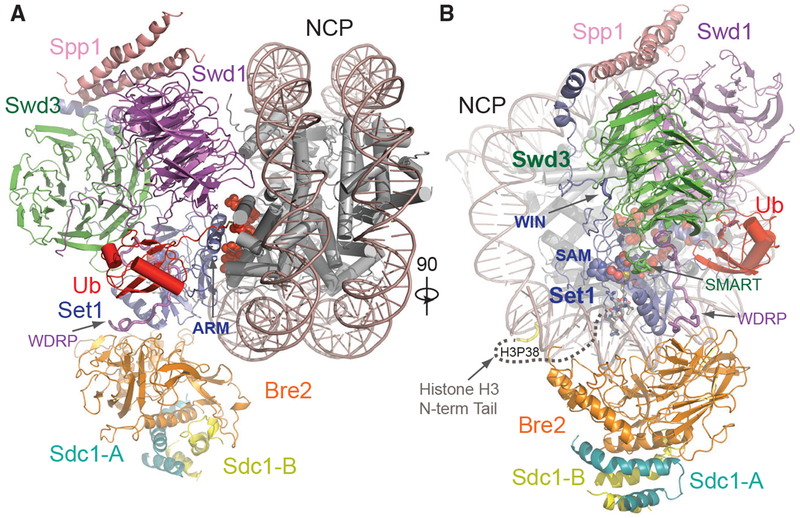
Figure 2. Overall Structure of the COMPASS eCM-uNCP Complex.
(A) Model of COMPASS-eCM bound to the H2Bub modified nucleosome. Set1 (blue), Swd1 (magenta), Swd3 (green), Bre2 (orange), Sdc1-A and B (turquoise and yellow), Spp1 (pink), and Ub (red) are shown in cartoon form. Nucleosomal DNA (dark salmon) is shown in cartoon form. The histone octamer is shown in cylinders. The SAM cofactor is shown in space-filling form. Residues forming the H2A-H2B acidic patch are shown in red spheres. The positions of the Swd1 WDRP and Set1 ARM helix are indicated by arrows.
(B) View of (A) rotated 90° and colored in the same scheme. The positions of the Set1 WIN motif (tube form), Swd3 SMART motif (tube), and Swd1 WDRP (tube) are indicated by arrows. A dashed line indicates the flexible histone H3 N-terminal tail spanning residue 9–38, which connects the first stretch of H3 seen in the NCP body (yellow tube) and the H3 (A1-R8) peptide (sticks) found in the Set1 SET domain.