Figure 3. DNA Binding Stabilizes COMPASS on Nucleosomes.
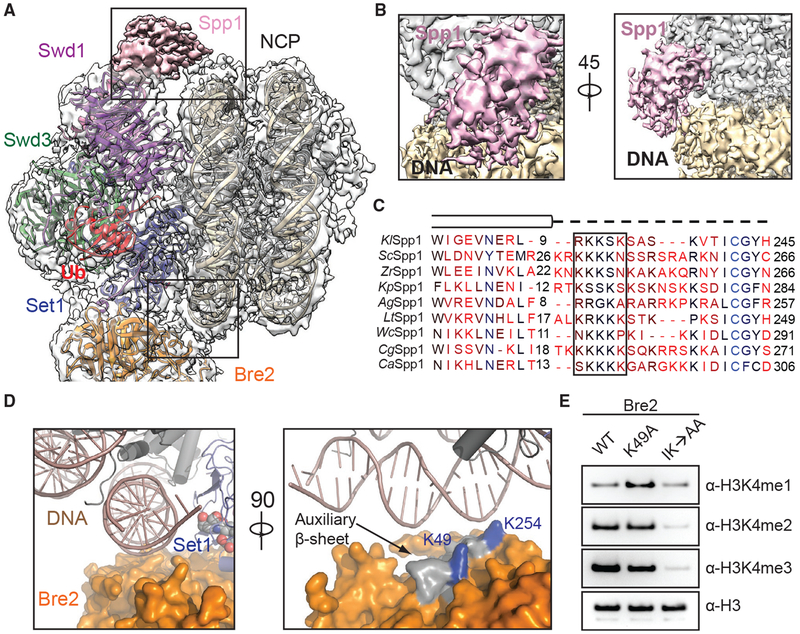
(A) Zoom-out view of the complex structure superimposed on the cryo-EM map. Major DNA-binding sites are boxed. Swd1 is shown in magenta, Swd3 in green, Set1 in blue, Bre2 in orange, Spp1 in pink, Ub in red, histones in dark gray, and nucleosomal DNA in wheat.
(B) Close-up views of the Spp1 DNA-binding domain density (pink) on nucleosomal DNA (wheat) in two different orientations.
(C) Structure-based sequence alignment of yeast Spp1 orthologs from K. lactis (Kl), S. cerevisiae (Sc), Z. rouxii (Zr), K. pastoris (Kp), A. gossypii (Ag), L. thermotolerans (Lt), W. ciferri (Wc), C. glabrata (Cg), and C. albicans (Ca). Cylinders denote α helices, while the dashed line indicates an unmodeled region, predicted to lack secondary structure. A basic region with potential DNA-binding function is boxed.
(D) Left: close-up view of Bre2 (orange in surface representation) looking down the nucleosomal DNA entry/exit point (cartoon representation, dark salmon). Right: 90° rotation of the left panel, highlighting the position of auxiliary β sheet (gray) on Bre2 relative to DNA. Two highly conserved lysine residues on the surface of this structural element are colored in blue.
(E) H3K4 methyltransferase activity of the COMPASS CM assembled with Bre2 mutants against nucleosomal substrates. The IK→AA double mutant of two conserved surface residues, Ile251 and Lys254, drastically reduces activity of the CM against H3K4.