Figure 7. The Set1 ARM Regulates COMPASS H2Bub Sensitivity.
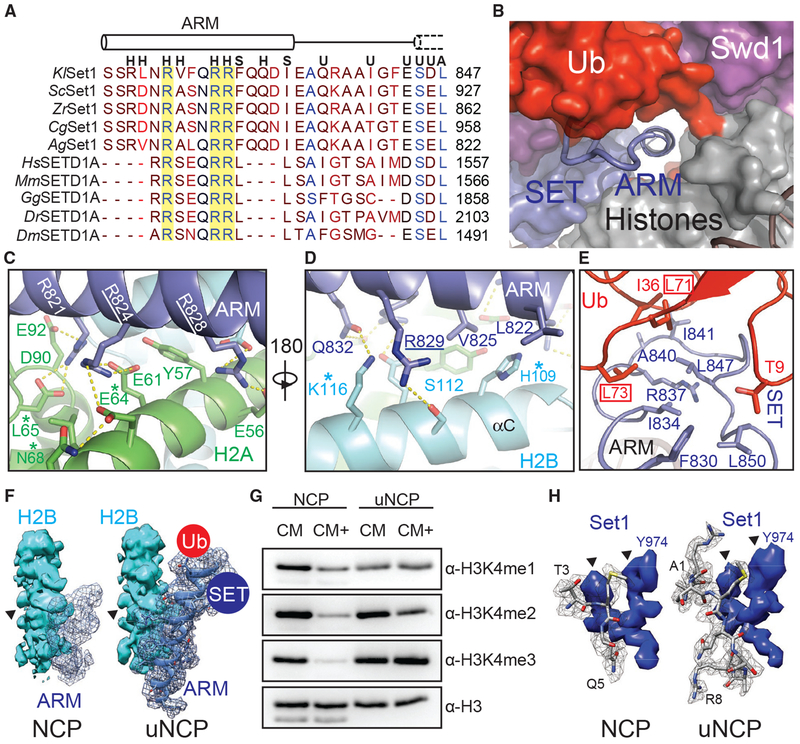
(A) Sequence alignment of the nSET ARM across K. lactis (Kl), S. cerevisiae (Sc), Z. rouxii (Zr), C. glabrata (Cg), A. gossypii (Ag), H. sapiens (Hs), M. musculus (Mm), G. gallus (Gg), D. rerio (Dr), and D. melanogaster (Dm) orthologs. The dotted cylinder represents the unraveled SET N-terminal helix. Cylinders represent helices, and lines indicate coiled structure. The strictly conserved arginine residues are highlighted by a yellow background. Letters above the alignment indicate the interactions the residue is involved in. (H) indicates a histone interaction, (S) indicates a SET domain interaction, (U) indicates a Ub interface, and (A) indicates a Sett ARM interaction.
(B) View of the tunnel formed by the junction between Set1 (blue, surface), Swd1 (magenta, surface), Ub (red, surface), and the histone octamer (gray, surface). Swd3 sits at the periphery (green, surface). The Set1 ARM helix (blue, cartoon) sits in the center of this tunnel. The loop connecting the SET domain and the ARM helix is shown in tube form.
(C) Close-up view of the Set1 ARM helix (blue) against the acidic surface of H2A (green). Side chains are shown in sticks. Two of the three strictly conserved arginine residues that impair H2Bub-H3K4me crosstalk (Kim et al., 2013) are underlined. H2A residues identified in (Nakanishi et al., 2008) critical for H2Bub-dependent H3K4 methylation are marked by an asterisk. Hydrogen bonds and salt bridges are shown as dashed yellow lines.
(D) Close-up view of the Set1 ARM (blue) against the H2B αC helix (cyan). Side chains are shown in sticks. The third strictly conserved arginine residue involved in the H2Bub-H3K4me crosstalk (Kim et al., 2013) is underlined. H2B residues identified by Nakanishi et al. (2008) to be important for H2Bub-dependent H3K4 methylation are marked by an asterisk. Hydrogen bonds and salt bridges are shown as dashed yellow lines.
(E) Close-up view of the three-way interface between the Set1 (blue) ARM, SET N terminus, and the Ub (red) Ile36 site. Side chains are shown in sticks. The ARM helix packs against the SET N terminus using an interlocking series of hydrophobic amino acids. Ub contributes to this interface by helping form the connecting loop between the ARM and SET regions through hydrophobic packing. Ub Leu71/Leu73, identified by Holt et al. (2015) to be important for H2Bub-stimulated H3K4me activity, are boxed.
(F) Comparison of the Set1 ARM density (blue mesh) between the unmodified NCP-bound (left) and uNCP-bound (right) structure of the COMPASS eCM. The neighboring H2B density (cyan, surface) was used to normalize the ARM densities. The key side-chain feature on H2B used for controlling the map contour level is indicated by a triangle. The uNCP density has the model of the ARM superposed in the mesh. The positions of where Ub and the SET domain dock on the ARM helix are indicated by a red and blue circle, respectively.
(G) H3K4 methyltransferase activity of the COMPASS CM and CM plus (CM+) on NCP and uNCP substrates. CM+ activity is suppressed relaive to the CM on NCPs and regains robust methyltransferase activity on uNCPs.
(H) Comparison of the H3 N-terminal tail density (gray mesh) at the Set1 catalytic cleft between the unmodified NCP-bound (left) and uNCP-bound (right) structure of the COMPASS eCM. The density of a nearby Set1 sequence was used to normalize the H3 N-terminal tail densities. The key features of the Set1 density for controlling the map contour level is indicated by triangles. The models of the H3 N-terminal tail centered at the H3K4 methylation site are shown in sticks.