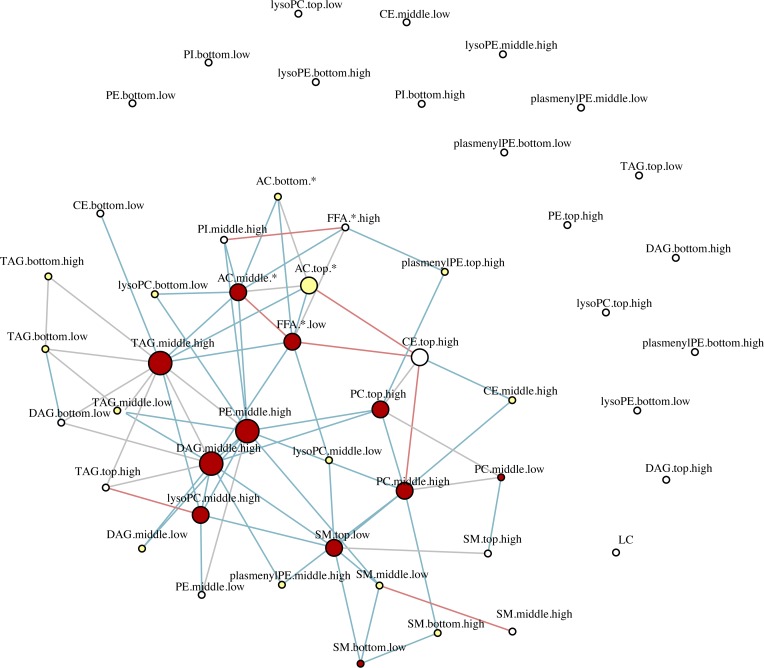
Figure 7. Differential network analysis.
To identify inter- and intraclass lipid correlates, we obtained the sparse partial correlation networks that captured the interdependencies between lipids. We used lipid grouping structure, obtained the superset of the network skeleton, and finally obtained the final stable network structures, the latter based on a bootstrapping method. Differential network analysis revealed differential loss of edges between various lipid classes in progressors characterized by 547 significant edges versus 1028 in nonprogressors (P < 0.0001) out of 55,460 possible permutations of bivariate correlations. The lines represent significant edges that were exclusively observed in nonprogressors (shown in blue) or progressors (shown in red). Common edges are shown in gray. The node size is proportional to the number of connectivity levels within and across lipid subclasses, and node colors represent number of cross-class connections (white, low; yellow, middle; red, high). Nodes are categorized by chain length (bottom, middle, top) and double bonds (low, high), with details shown in Supplemental Table 3.