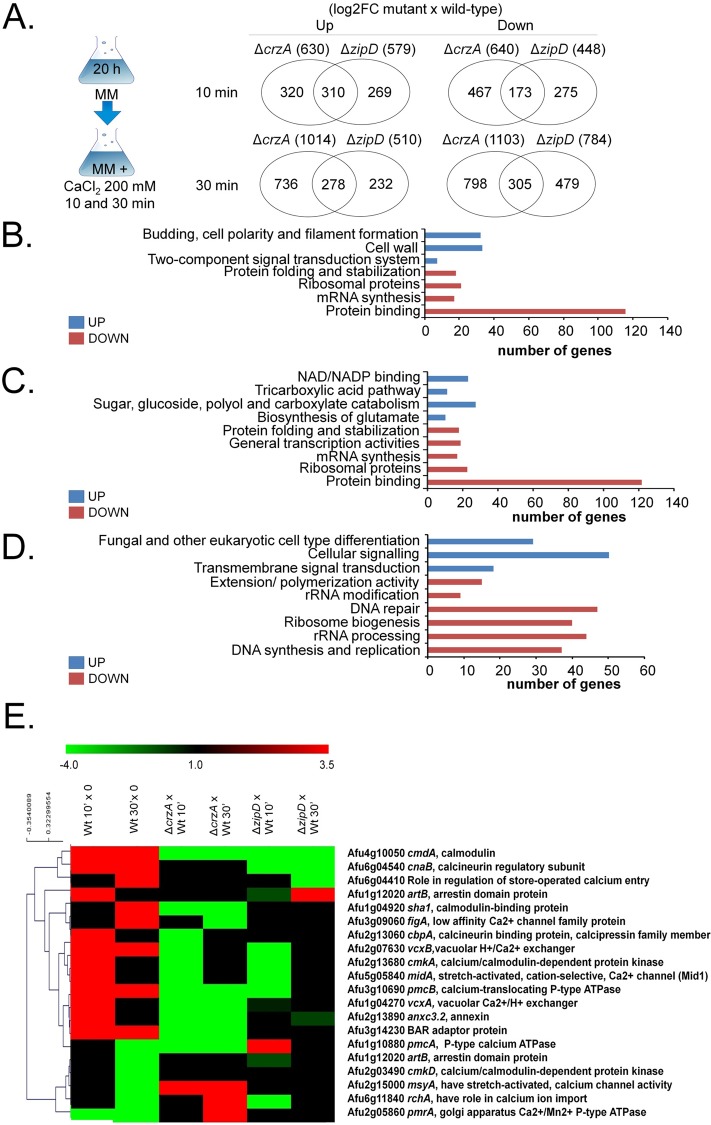
Fig 3. The ΔcrzA and ΔzipD show common and distinct transcriptional profiles.
(A) Venn diagram comparing the genes specifically expressed in each mutant relative to the wild-type strain. (B) A summary of the FunCat terms over-represented up or down regulated (adjusted p-value < 0.05) in log2FC ΔzipD versus ΔcrzA post transfer to 200 mM CaCl2 for 10 and 30 mins. For the full list refer to S3–S8 Tables. (C) A summary of the FunCat terms over-represented up or down regulated (adjusted p-value < 0.05) but uniquely expressed in ΔzipD in log2FC ΔzipD versus wild-type post transfer to 200 mM CaCl2 for 10 and 30 mins. For the full list refer to S2–S8 Tables. (D) A summary of the FunCat terms over-represented up or down regulated (adjusted p-value < 0.05) but uniquely expressed in ΔcrzA in log2FC ΔzipD versus wild-type post transfer to 200 mM CaCl2 for 10 and 30 mins. For the full list refer to S2–S8 Tables. (E) The RNA-seq data for annotated genes corresponding to the expression of genes encoding selected proteins putatively involved in calcium metabolism. The wild-type is shown as 10 and 30 min calcium stress versus time zero (20 hours growth), and gene deletion strains are shown as the deletion strain versus the equivalent wild-type 10 and 30 min time points (the mutant values have been normalised to the basal level of expression of each gene before stress, i.e., expression ratios are being compared: wild-type 10 min versus time zero divided by a specific mutant 10 and 30 min versus time zero). Hierarchical clustering was performed in MeV (http://mev.tm4.org/), using Pearson correlation with complete linkage clustering.