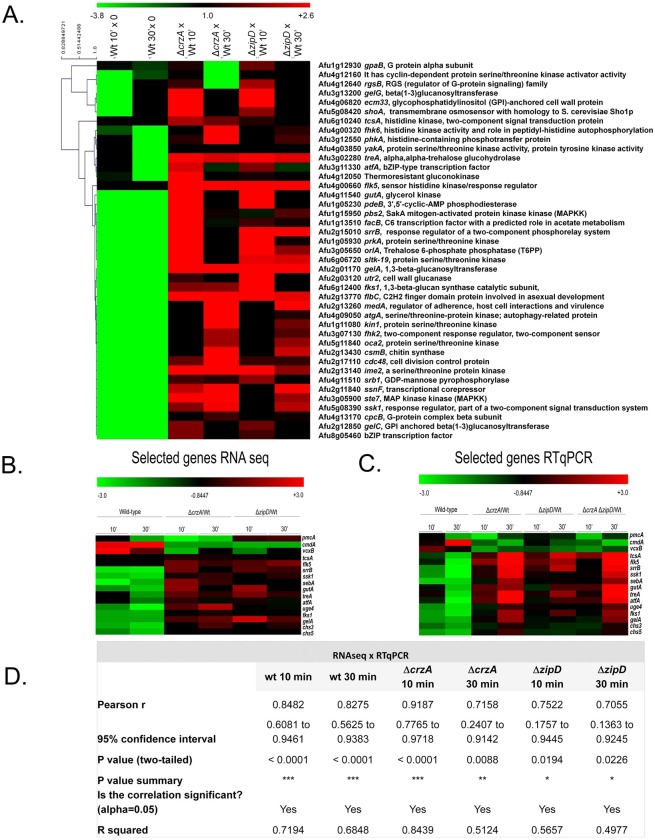
Fig 4. The ΔcrzA and ΔzipD influence calcium, osmotic stress and signaling, and cell wall metabolism.
The RNA-seq data for annotated genes corresponding to the expression of genes encoding selected proteins putatively involved in calcium metabolism (A) Selected proteins putatively involved in calcium signal transduction, osmotic stress, cell wall biosynthesis, and transcription factors.The wild-type is shown as 10 and 30 min calcium stress versus time zero (20 hours growth), and gene deletion strains are shown as the deletion strain versus the equivalent wild-type 10 and 30 min time points (the mutant values have been normalised to the basal level of expression of each gene before stress, i.e., expression ratios are being compared: wild-type 10 min versus time zero divided by a specific mutant 10 and 30 min versus time zero). Hierarchical clustering was performed in MeV (http://mev.tm4.org/), using Pearson correlation with complete linkage clustering. (B) Heatmap for the RNAseq values of sixteen selected genes involved in calcium metabolism, osmotic stress, and cell wall metabolism. (C) Heat map of RTqPCR for the same sixteen selected genes from (B). The wild-type, ΔcrzA, ΔzipD, and ΔcrzA ΔzipD mutant strains were grown for 20 h at 37°C and transferred to 200 mM CaCl2 for 0, 10, and 30 mins. Gene expression was normalized using cofA (Afu5g10570). Standard deviations present the average of three independent biological repetitions (each with 2 technical repetitions). (D) The expression of these sixteen genes as measured by RT-qPCR showed a high level of correlation with the RNA-seq data (Pearson correlation from 0.7055 to 0.9187).