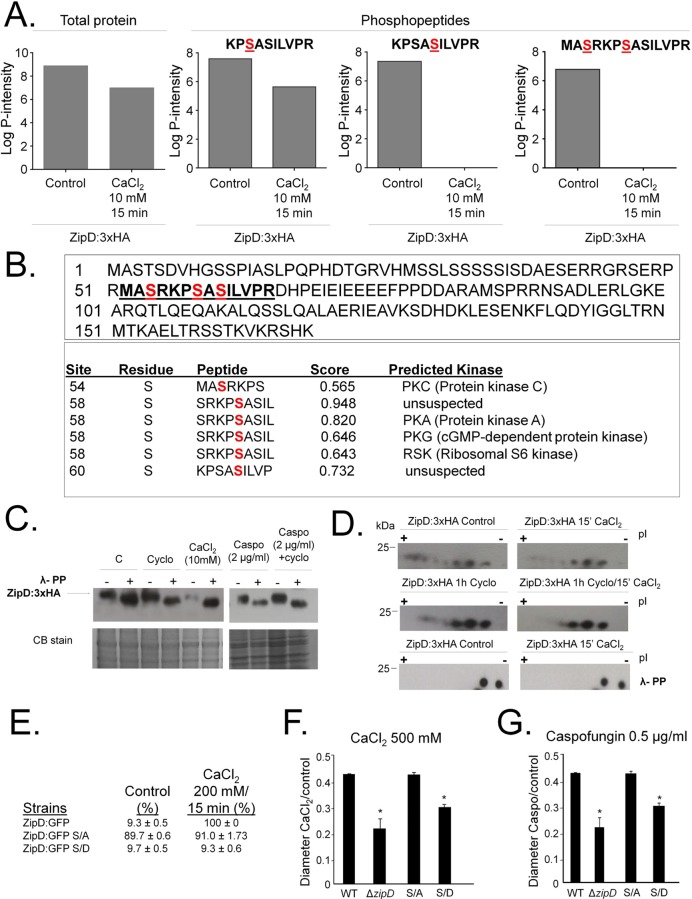
Fig 6. A. fumigatus ZipD:3xHA is dephosphorylated upon calcium stress but not by calcineurin.
(A) The ZipD amino acid sequence. The underlined peptide shows the serine residues (in red) that were identified as phosphorylated. NetPhos 3.1 (http://www.cbs.dtu.dk/services/NetPhos/) predicted the serine residues to be phosphorylated by PKC (Protein kinase C), PKA (Protein kinase A), PKG (cGMP-dependent protein kinase), and RSK (Ribosomal S6 kinase). (B) Relative abundance of the identified phosphopeptides. (C) Immunoblot analysis for immunoprecipated ZipD:3xHA. The A. fumigatus ZipD:3xHA strain was grown for 24 h at 37°C (C = Control) and transferred to either cyclosporin (Cyclo) 9 μg/ml for 1 h, CaCl2 10mM for 15 mins, or caspofungin 2.0 μg/ml for 1 h, or cyclosporin (9 μg/ml) for 1 h followed by 2.0 μg/ml caspofungin for another additional 1 h. Anti-HA antibody directed against the HA epitope was used to detect the ZipD:3xHA. The immunoprecipitated ZipD:3xHA was treated (+) or not (-) with Lambda phosphatase for 1 h at 30°C. A Coomassie Brilliant Blue (CBB)-stained gel is shown as an additional loading control. (D) Immunoprecipated ZipD:3xHA (same experimental design from C) was subjected to two-dimensional gel electrophoresis coupled with Western blotting. Anti-HA antibody directed against the HA epitope was used to detect the ZipD:3xHA. The immunoprecipitated ZipD:3xHA was treated (+) or not (-) with Lambda phosphatase for 1 h at 30°C. (E) Percentage of ZipD:GFP translocated to the nuclei in ZipD:GFP, ZipD:GFP S/A, and ZipD:GFP S/D strains. (F and G) Growth phenotypes of ZipD:GFP, ZipD:GFP S/A, and ZipD:GFP S/D on CaCl2 and caspofungin. The strains were grown for 5 days at 37°C.