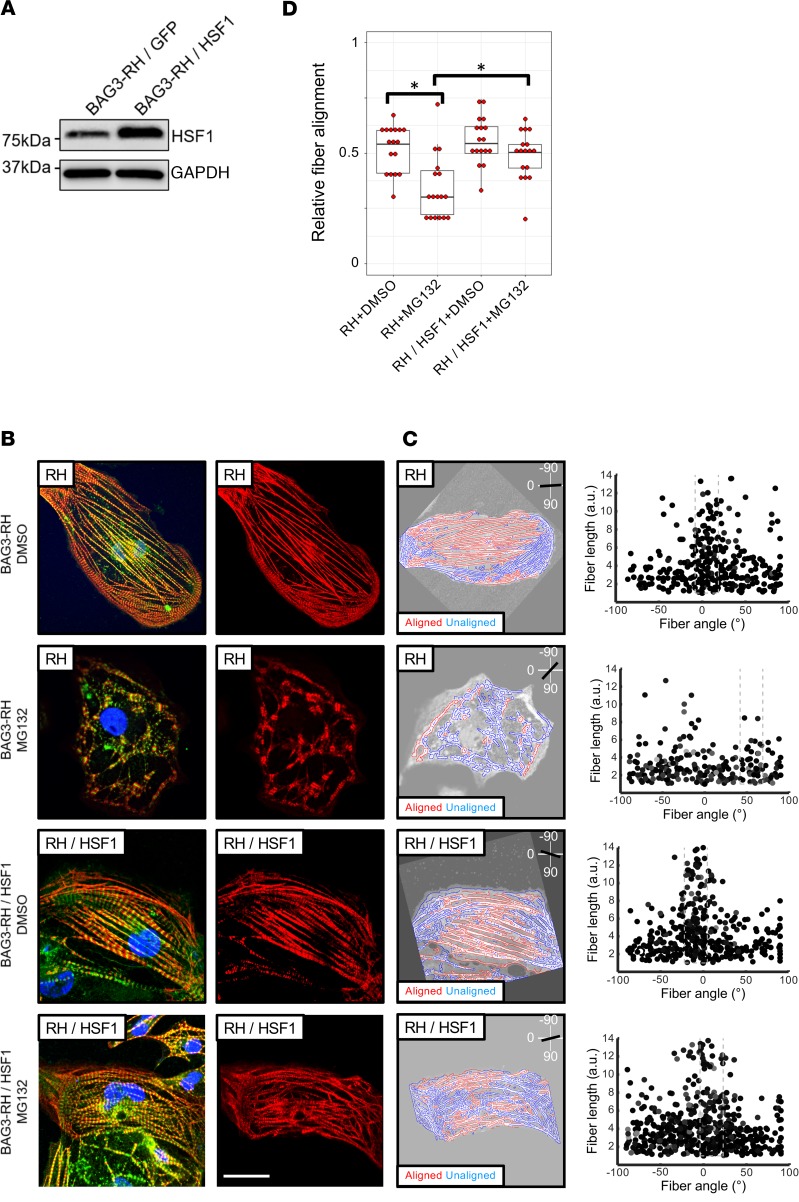
Figure 5. Overexpression of HSF1 mitigates proteasome inhibition–induced myofibrillar disarray in induced pluripotent stem cell–derived cardiomyocytes (iPCS-CMs) with the BAG3-R477H (RH) mutation.
(A) Western blot for HSF1 and GAPDH (loading control) in BAG3-RH iPCS-CMs transduced with HSF1 and GFP lentivirus. (B) Visualization of myofibrillar organization in untransduced and HSF1-transduced BAG3-RH iPSC-CMs following treatment with MG132 (25 μM, 15 hours) or DMSO. Red, cardiac troponin T; green, α-actinin; blue, DAPI. Images display representative cells. Right panels show cardiac troponin channel only for clarity. Scale bar: 20 μm. (C) Quantification of myofibrillar disorganization/disorganization in cells in B. Left panels show individual myofiber boundaries (identified from cardiac troponin channel) defined using an edge detection algorithm in MATLAB (see Methods) and are displayed as red or blue if aligned or unaligned, respectively, with the predicted long axis of the parent cell, indicated in the top right corner. Right panels display each myofiber from the corresponding parent cell (left panels) as a single point and are plotted according to their length (y axis) and angle relative to the predicted long axis (x axis) of the parent cell. The dashed box represents ±15° of the predicted long axis of the cell. (D) Relative myofiber alignment based on edge detection measurements in untransduced BAG3-RH iPSC-CMs and BAG3-RH iPSC-CMs overexpressing HSF1 following addition of MG132 (25 μM, 15 hours) or DMSO. Each data point represents average myofiber alignment from a single cell. For each condition, 17 randomly selected cells per analyzed. Boxplots show median and interquartile range (IQR); whiskers extend 1.5 times the IQR. *P < 0.05 (unpaired 2-tailed t test) following Bonferroni correction for multiple comparisons. Data is representative of 3 independent experiments.