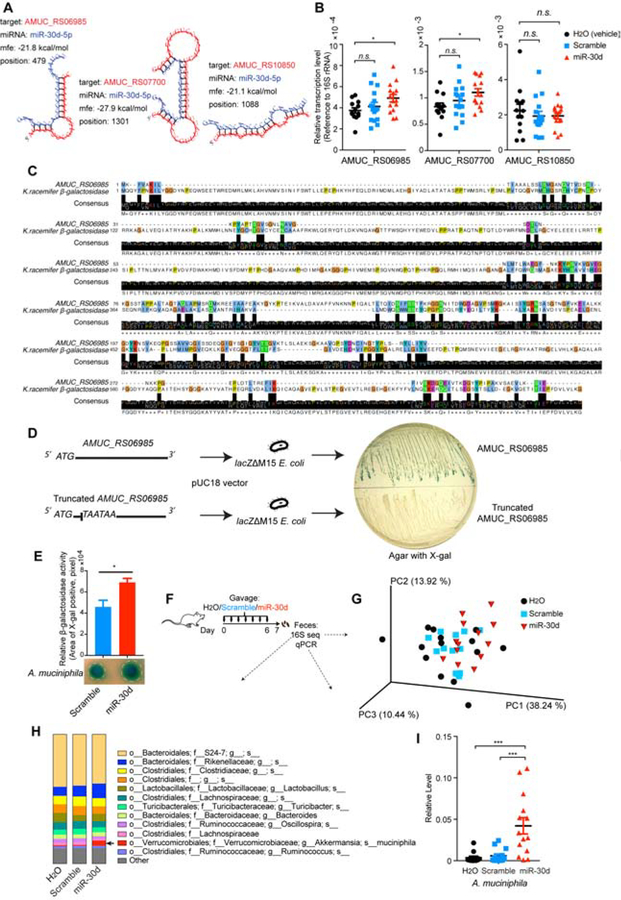
Figure 5. miR-30d Enhances β-galactosidase of A. muciniphila and Expands A. muciniphila in vivo.
(A) A. muciniphila genes (AMUC_RS06985, AMUC_RS07700, AMUC_RS10850) were predicted to be targeted by miR-30d by sequence blast and predicted for secondary structure property (minimum free energy, mfe) by RNAhybrid. (B) A. muciniphila was grown in the presence of synthetic miR-30d, scramble or H2O (vehicle). Transcripts of the predicted targeting genes at logarithmic phase were quantified by qPCR normalized to 16S rRNA. H2O (vehicle) n=13, scramble n=15, miR-30d n=15, Error bars denote mean ± SEM, one-way ANOVA Dunnett’s multiple comparisons test. (C) Alignment protein sequence of AMUC_RS06985 of A. muciniphila and β-galactosidase of Ktedonobacter racemifer (K. racemifer). (D) AMUC_RS06985 of A. muciniphila or its truncated sequence was cloned into a β-galactosidase-deficient (lacZΔM15) E. coli. The cloned E. coli colonies were grown on an X-gal-containing agar. (E) A. muciniphila was grown on BHI agar containing lactose and β-galactosidase activity indicator X-gal, and was treated with synthetic miR-30d or scramble. β-galactosidase activity was quantified according to the color change, n=5 each group, Error bars denote mean ± SEM, paired t test. (F-I) The effect of oral administration of synthetic miR-30d on the gut microbiome. Mice were immunized with MOG and orally gavaged with 250 pmol synthetic miR-30d, scramble or H2O (vehicle) for 7 days. Feces were collected at day 7 and bacterial 16S rDNA sequence-based microbiome surveys were performed. (F) Experimental scheme. (G) Principal coordinates analysis (PCoA) based on weighted UniFrac metrics and (H) Relative abundance of bacteria by 16S sequencing was classified at a species-level taxonomy. H2O (vehicle) n=14, scramble n=13, miR-30d n=13. One-way ANOVA Dunnett’s multiple comparisons test. Arrow identifies species that were significantly higher in miR-30d group compared to the other two groups. H2O (vehicle) vs miR-30d P= 0.0129, scramble vs miR-30d P= 0.0087. (I) qPCR quantification of the relative abundance of A. muciniphila by measuring the 16S rDNA gene, referenced to universal 16S rDNA. H2O (vehicle) n=14, scramble n=14, miR-30d n=13. Error bars denote mean ± SEM, One-way ANOVA Tukey’s multiple comparisons test.