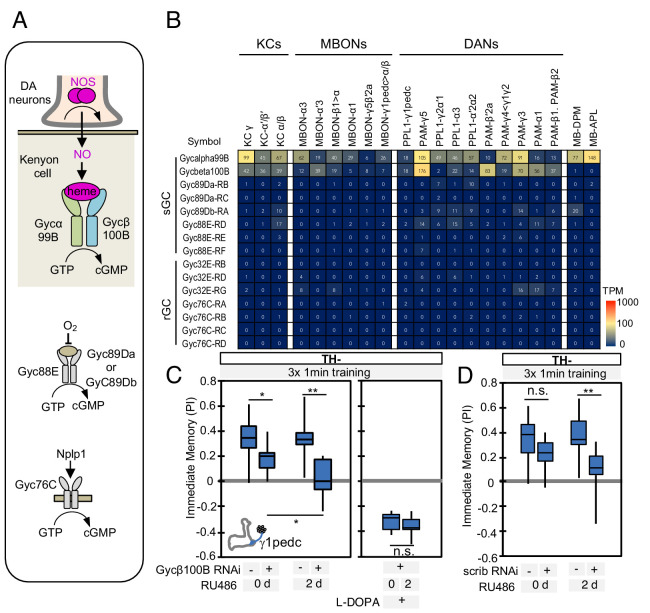
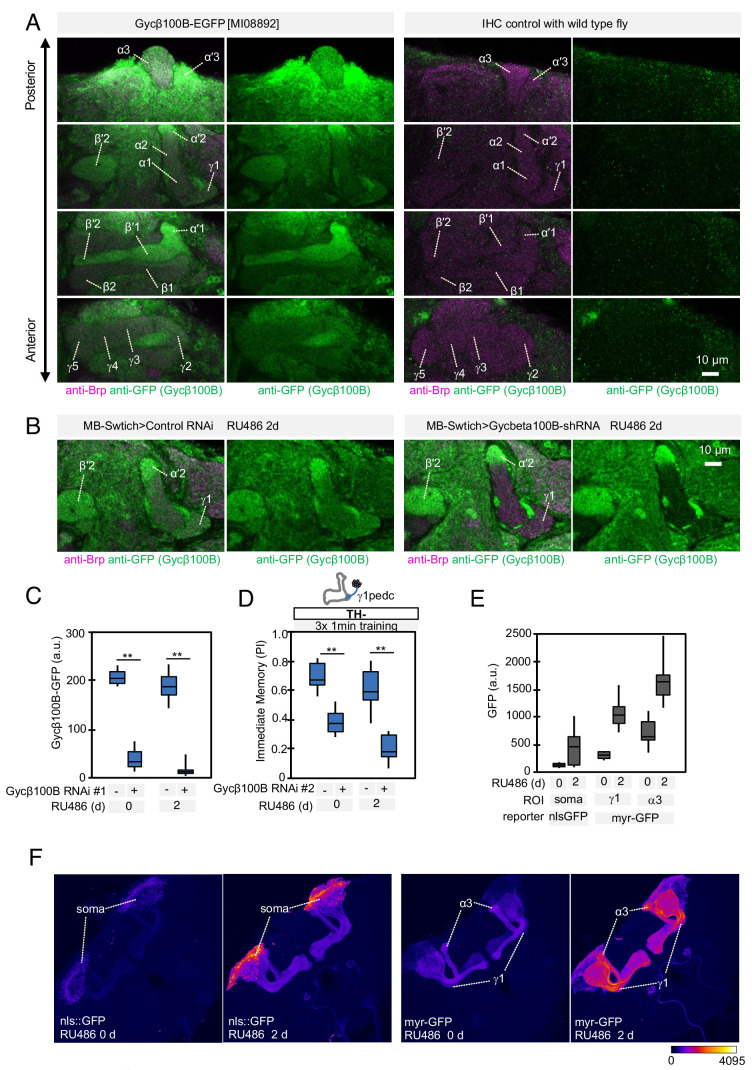
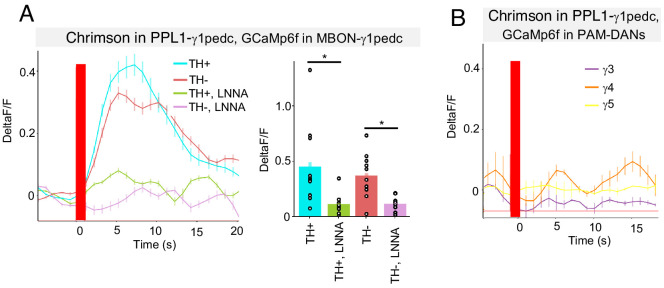
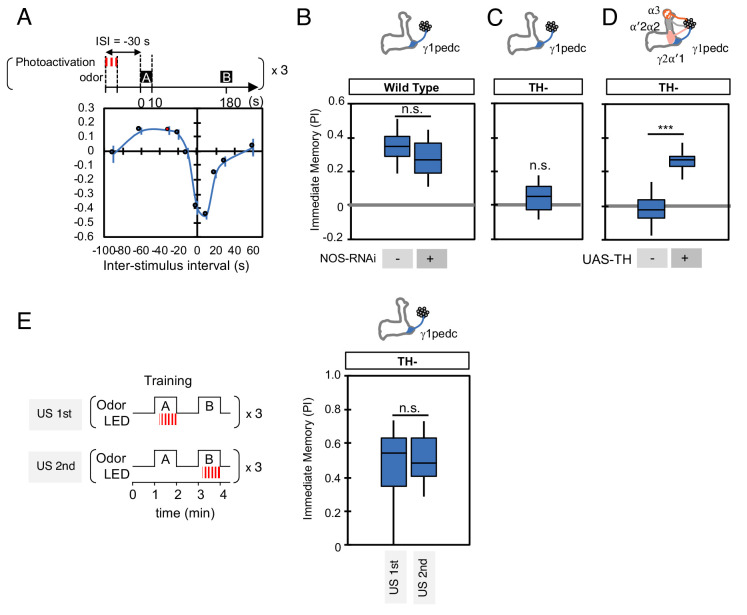
Figure 5. Soluble guanylate cyclase in the KCs is required to form NO-dependent memory.
(A) Diagram of soluble or receptor guanylyl cyclases in Drosophila. (B) RNA-seq data indicate coexpression of Gycα99B and Gycβ100B in KCs, MBONs and DANs. For comparison, the expression levels of other guanylate cyclase genes are also shown. Note that RNA-Seq detected transcripts of neuropeptide gene Nplp1 in both PPL1-γ1pedc and PAM-γ5 (Figure 3—figure supplement 8), but expression of its receptor Gyc76C was barely detectable compared to Gycα99B and Gycβ100B. (C) Induction of Gycβ100B-shRNA in Kenyon cells by activating MB247-switch driver (Mao et al., 2004) with RU-486 feeding reduced the positive-valence memory induced by PPL1-γ1pedc. We also observed a partial effect in the flies without RU-486, presumably due to leaky expression (Figure 5—figure supplement 1E and F). Negative-valence memory with additional feeding of L-DOPA and carbidopa was not affected by Gycβ100B-shRNA induction in KCs. Memories immediately after 3 × 1 min training are shown. The bottom and top of each box represents the first and third quartile, and the horizontal line dividing the box is the median. (D) Induction of scrib-shRNA in KCs also reduced the positive-valence memory induced by activation of PPL1-γ1pedc in a TH mutant background. The whiskers represent the minimum and maximum. N = 12–16. Asterisk indicates significance of designated pair: *, p<0.05; **,p<0.01; n.s., not significant.