Figure 2.
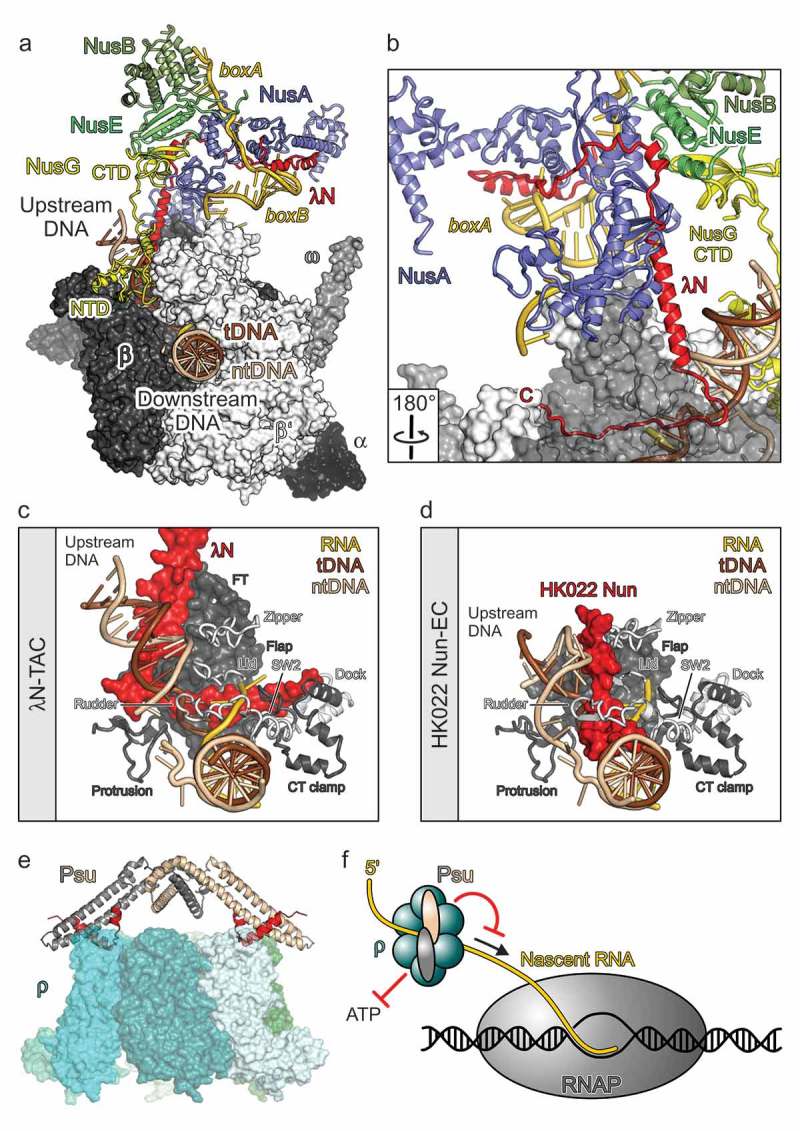
Molecular models of phage factors modulating host transcription elongation and termination.
(a) A Single-particle cryoEM structure of an λN-TAC [17]. RNAP subunits in surface representation. Nucleic acids, Nus factors, and λN in cartoon representation. (b) λN remains highly extended in the λN-TAC, allowing short peptides along its sequence to interact with spatially widely distributed regions on nascent RNA, Nus factors and RNAP. RNAP subunit β as semi-transparent surface. Rotation symbol – view relative to (a). (c) Interaction of the C-terminal region of λN with nucleic acids and various elements of RNAP in and around the active site cleft (β elements: flap, FT – flap tip, protrusion, CT clamp – C-terminal clamp; β’ elements: zipper, lid, rudder, SW2 – switch 2, dock). View as in (a). (d) HK022 Nun interacting with nucleic acids and RNAP elements [60]. Same orientation of RNAP as in (d). (e) presumed mode of action of Psu [16]. By inhibiting ρ’s ATPase, Psu will hinder the translocation of ρ along the nascent RNA toward RNAP (arrow). Red symbols – inhibition. (f) Docking model of the phage P4 Psu protein interacting with E. coli transcription termination factor ρ [62]. (c) and (d) adapted from [17].
