Figure 3.
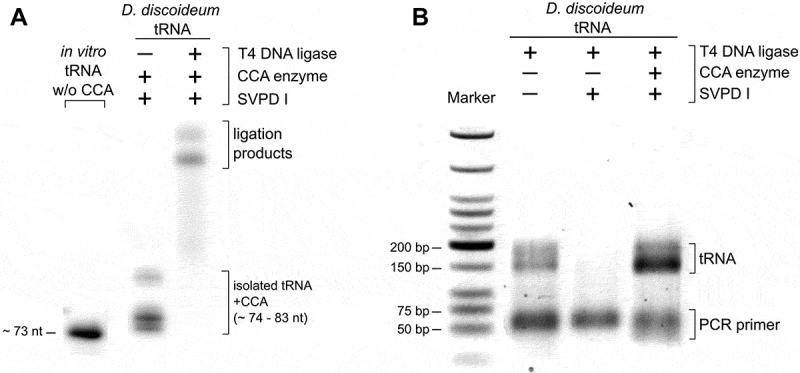
Hairpin adapter ligation to a tRNA pool preparation. (A) To introduce a radioactive label for visualization, CCA ends of the D. discoideum tRNA pool were removed by snake venom phosphodiesterase and restored by CCA-adding enzyme (D. discoideum) and NTPs spiked with α-32P-ATP. T4 DNA ligase fused the labelled tRNA pool to the hairpin adapter at high efficiency. (B) After reverse transcription and ligation of the second adapter, cDNA was amplified with indexed primers for Illumina deep sequencing. The original tRNA pool of D. discoideum as well as pool samples without or with restored CCA ends were analysed. Only samples with CCA ends (original or restored) gave rise to amplification products. The product length of 150 to 200 bp corresponds to the expected size of PCR products consisting of adapters and complete or partial tRNA sequences.
