Fig. 4. Single-molecule visualization of DNA-PK removal by MRN and CtIP.
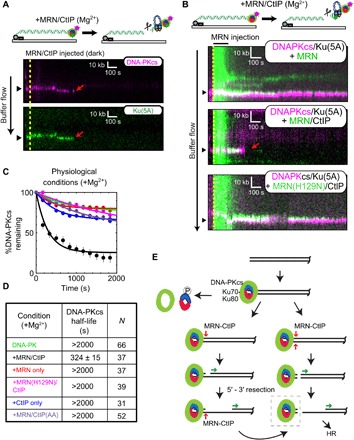
(A) Illustration and kymographs of DNA-PKcs (magenta) and Ku (green) upon injection of MRN and CtIP [both unlabeled (dark)]. (B) Kymographs of DNA-PKcs (magenta) with Ku (unlabeled), with injection of MRN (green) alone (top), or MRN with CtIP (middle), or a nuclease-dead mutant of MRN (H129N) with CtIP (bottom). (C and D) Lifetimes and associated half-lives of the DNA-PK complex (as observed by DNA-PKcs occupancy) upon no injection (green), injection with MRN and CtIP (black), MRN alone (red), nuclease-deficient H129N MRN and CtIP (magenta), CtIP only (blue), or MRN with phospho-blocking T847A/T859A CtIP (purple). Table shows half-life of DNA-PKcs under various conditions and the number of molecules observed (N). (E) Schematic model for DNA-PK removal from DSB ends, as discussed in the text. DNA-PKcs binds to the Ku heterodimer bound to DNA at the DSB. Transition from NHEJ to homologous recombination (HR) results from (i) DNA-PKcs phosphorylation of Ku, resulting in dissociation of Ku as well as DNA-PKcs from ends, (ii) DNA-bound DNA-PK stimulation of MRN single-strand endonucleolytic cleavage followed by 5′ to 3′ resection, or (iii) DNA-PK stimulation of MRN double-stranded endonucleolytic cleavage resulting in DNA-PK loss and 5′ to 3′ resection. Long-range 5′ to 3′ resection ensues from the nick or a new DSB, creating 3′ single-stranded DNA that is used for homology search. The DNA-bound DNA-PK complex (dashed box) is the species isolated in the modified ChIP protocol (Fig. 5 and fig. S4).
