Fig. 5. DNA-PK–associated DNA end fragments are observed at AsiSI breaks in human cells using GLASS-ChIP.
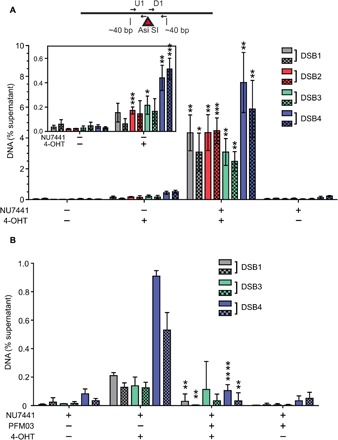
(A) Small dsDNA products resulting from nucleolytic cleavage of DNA-PK–bound AsiSI–generated DNA ends (dashed line box, Fig. 4E) were isolated from U2OS cells treated with 4-OHT or vehicle for 4 hours as indicated. NU7441 (10 μM) was added as indicated for 5 hours starting at 1 hour before 4-OHT addition to induce AsiSI. DNA-PK–bound DNA was isolated using a modified ChIP protocol (GLASS-ChIP, fig. S4) and quantified by qPCR using primers located ~30 nt from the AsiSI cut site (results from primers ~300 nt from cut sites in fig. S5). Primer set U1 (solid) is upstream, whereas D1 (checkered) is downstream of the AsiSI cut sites. The DNA quantitated from U2OS cells in the presence or absence of 4-OHT and a DNA-PKcs inhibitor (NU7441) is shown for each AsiSI site (46). The inset magnifies the results for experiments performed in the absence of NU7441. Results are from three independent biological replicates, with Student’s two-tailed t test performed; *P < 0.05, ** P < 0.01, ***P < 0.001, in comparison to equivalent samples without 4-OHT. (B) The GLASS-ChIP protocol was performed as in (A) using cells treated with a DNA-PKcs inhibitor (NU7441, 10 μM), a Mre11 inhibitor (PFM03, 100 μM), and 4-OHT for 1 hour as indicated. Results are from three independent biological replicates, with Student’s two-tailed t test performed; **P < 0.005 and **** P < 0.0001, in comparison to equivalent samples without PFM03.
