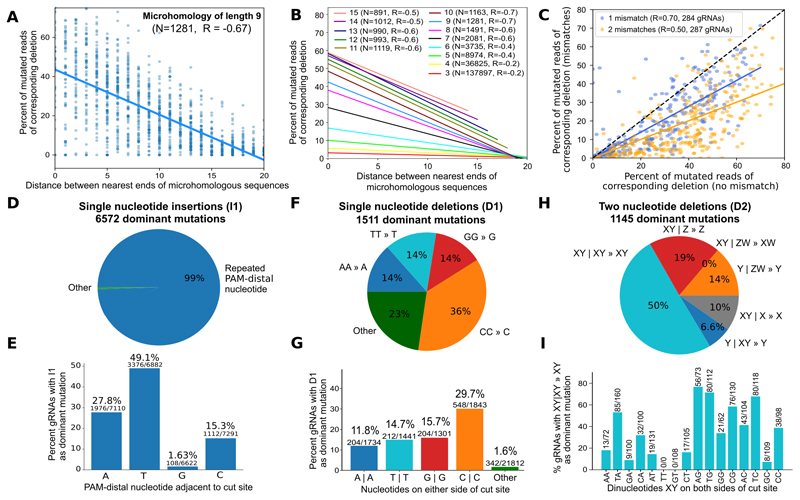
Figure 4. Local sequence context strongly influences editing outcomes in the explorative set of gRNA-target pairs.
A. Nearby matching sequences are used as substrate for microhomology-mediated repair more frequently than distant ones. Fraction of mutated reads (y-axis) for increasing distance between 1,281 matching sequences of length 9 (x-axis) (blue markers) in K562 cells, and a linear regression fit to the trend (solid line; Pearson's R=-0.67). Reproducibility of measurements is presented in Figure 5C.
B. Frequency of microhomology-mediated repair depends on the length of and distance between the matching sequences. Same as (A), but linear regression fits only for microhomologies of lengths 3 (red, bottom) to 15 (pink, top), with the number of pairs of matching sequences considered (N) and Pearson’s correlation (R) noted in the legend.
C. Mutations in microhomology sequence reduce repair outcome frequency, but corresponding deletions are still present. The fraction of mutated reads associated with the particular microhomology with mismatches (y-axis) vs without mismatches (x-axis) stratified by the number of mismatches (blue: one mismatch, yellow: two mismatches). Solid lines: linear regression fits; dashed black line: y=x; Pearson's R provided in legend.
D. Single nucleotide insertions are only dominant when repeating the PAM-distal nucleotide. Percentage of the 6,572 gRNAs for which insertion of a specific nucleotide is most frequent in all replicates (“dominant allele”; area of wedge) stratified by whether the PAM-distal nucleotide adjacent to the cut site is inserted (blue), vs. all other outcomes (green).
E. Insertions of thymine dominate often, while guanines are rarely inserted with reproducibly high frequency. The percentage of gRNAs that have a dominant single nucleotide insertion (y-axis), stratified by their PAM-distal nucleotide at the cut site (x-axis).
F. Dominant single nucleotide deletions usually remove one nucleotide from a repeating pair at the cut site. Percentage of the 1,511 gRNAs with a dominant single nucleotide deletion (area of wedge) of a repeating A (blue), repeating T (teal), repeating G (red), repeating C (orange), or a base from a non-repeat (green).
G. Dominance of single nucleotide deletions depends on both bases adjacent to the cut site. The percentage of gRNAs that have a dominant single nucleotide deletion (y-axis), stratified by the two bases on either side of the cut site (x-axis).
H. Two nucleotide deletions that are dominant favour repeats. Percentage of the 1,145 gRNAs with a dominant size two deletion (area of wedge) that delete a repeat (XY | XY » XY, teal), delete PAM-distal nucleotides (XY | Z » Z, red), delete one PAM-distal and one PAM-proximal nucleotide (XY | ZW » XW, purple), delete PAM-proximal nucleotides (Y | ZW » Y, orange), delete a PAM-distal nucleotide flanked by a repeating base (XY | X » X, grey), or delete a PAM-proximal nucleotide flanked by a repeating base (Y | XY » Y, blue). X, Y, Z, W - any nucleotide; | - cut site.
I. PAM-distal guanine at the cut site promotes, while PAM-distal thymine at the cut site demotes the frequency of dominant dinucleotide repeat contraction. The percentage of gRNAs with a dinucleotide repeat that have the corresponding dominant two nucleotide deletion (y-axis), stratified by the two bases in the repeated sequence (x-axis).