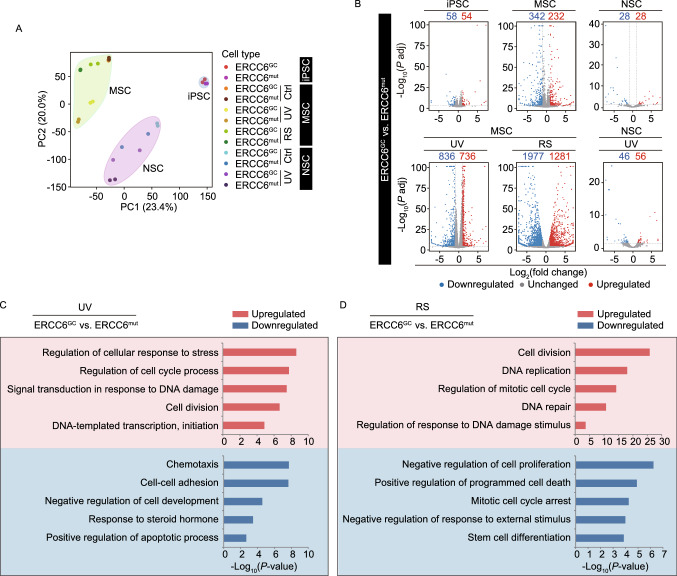
Figure 6.
The global gene expression profiles of CS-iPSCs and gene-corrected CS-iPSCs and their adult stem cell derivatives. (A) PCA of CS cells and GC cells in the absence or presence of UV (Ctrl or UV), as well as under replicative senescence (RS) stress. Each point represents a sample. Data points were computed based on Log2(FPKM + 1). (B) Volcano plots showing the differentially expressed genes between CS-iPSCs and GC-iPSCs, between CS-MSCs and GC-MSCs, and between CS-NSCs and GC-NSCs in the absence of UV (the upper panel) or in the presence of UV (the lower panel, UV), or under RS stress (the lower panel, RS). Red represents upregulated genes, and blue represents downregulated genes. (C) Gene Ontology Biological Process (GO-BP) enrichment analysis of significantly upregulated/downregulated genes in GC-MSCs compared to CS-MSCs upon UV treatment. Red represents upregulated genes, and blue represents downregulated genes. (D) Gene Ontology Biological Process (GO-BP) enrichment analysis of significantly upregulated/downregulated genes in GC-MSCs compared to CS-MSCs under RS stress. Red represents upregulated genes, and blue represents downregulated genes