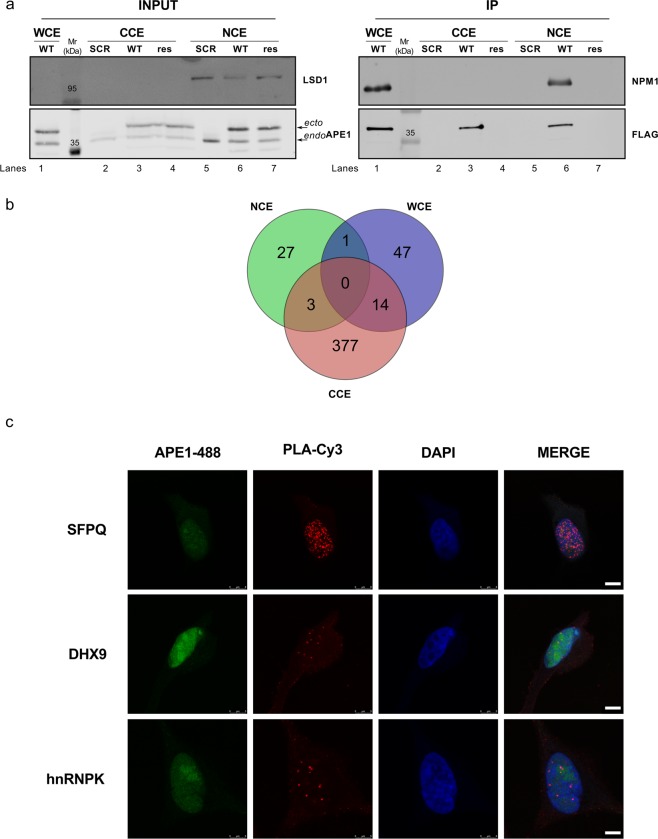
Figure 2.
Proteomic characterization of the APE1 interactome. (A) Representative Western blotting to confirm APE1 pulldown in the co-immunoprecipitation experiment. Western blotting analysis was performed on total HeLa cell clone extracts (INPUT) and on co-immunoprecipitated material (IP) with specific antibodies for APE1 and FLAG. The endogenous (endo) and ectopic (ecto) form of the APE1 protein is visible. The resulting material was tested for the occurrence of NPM1, a known APE1 interactor. LSD1 was used to probe nuclear enrichment. SCR, HeLa cell clone transfected with empty vector; WT, HeLa cell clone expressing APE1-FLAG tagged protein; res, co-immunoprecipitated with a resin lacking the FLAG antibody; WCE, whole cell extract; NCE, nuclear cell extract; CCE, cytoplasmic cell extract. (B) Venn diagram showing APE1-interacting partners identified in whole-cell lysates (WCE), or nuclear (NCE) and cytoplasmatic (CCE) extracts. (C) Nucleoplasmic interaction between APE1 and three identified interactors. HeLa cells were seeded on a glass coverslip and the PLA reaction was carried out using anti-APE1, anti-SFPQ, anti-DHX9 and anti-hnRNPK antibodies. APE1 localization was detected by using an anti-APE1 antibody and visualized in green. Confocal microscopy analysis highlighted the presence of distinct fluorescent red dots (PLA signals) indicating the occurrence of in vivo interaction between APE1 and its protein partners. DAPI staining was used as a reference for the nuclei. See also Supplementary Figs. S1 and S2 for negative controls. Bars, 8 µM.