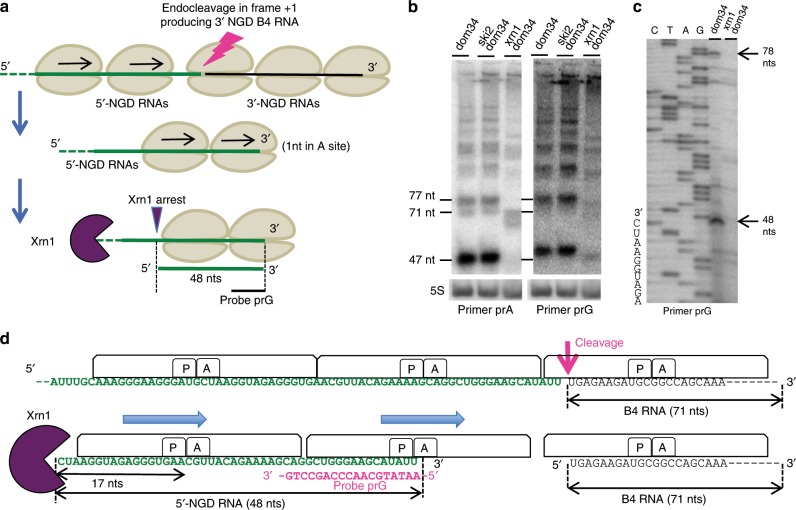
Fig. 4. Analysis of the fate of 5′-NGD RNAs.
a Schematic model of mRNA1 before and after the endonucleolytic cleavages producing B4 RNAs. 5′-NGD resulting RNAs are shown here covered by two ribosomes and processed by Xrn1 to 48-nt RNAs (see also Supplementary Fig. 4 for 5′-NGD RNAs covered by three ribosomes). b 8% PAGE followed by northern blot analysis using probe prG showing steady-state levels of RNAs in the indicated mutant strains (left panel). Same membrane has been probed with prA as a ladder (right panel), and sizes of B5 (77nt), B4 (71 nt) and B1 (47nt) are indicated. The 5S rRNA served as a loading control. c Primer extension experiments using probe prG to determine the 5’-end of RNAs (see also Supplementary Fig. 5). d Schematic model of ribosome positioning on mRNA1 before and after the unique endonucleolytic cleavage producing B4 RNAs, localized 8 nts upstream of the first P-site nt. The position of disomes on the resulting 48-nt 5’-NGD RNA is shown with the distal ribosome having 1 nt in the A site (see also Supplementary Fig. 4). Source data are provided as a Source Data file.