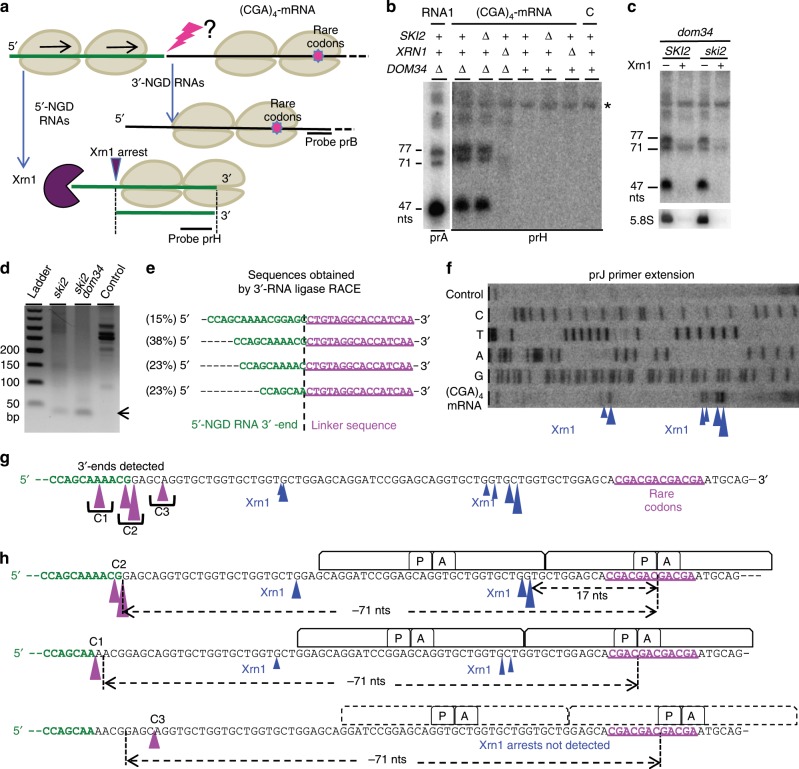
Fig. 6. Identification of the endonucleolytic cuts on the NGD targeted (CGA)4-mRNA.
a Schematic view of the (CGA)4-mRNA. 5′- and 3′-NGD RNAs are products of NGD. The lightning flash represents the potential endonucleolytic cleavage upstream of the ribosome stall site. Probes prB and prH are indicated. 5′-NDG RNAs are shown processed by Xrn1 as described in Figs. 4a, d and Supplementary Fig. 4. b Northern blot analysis using probe prH showing steady-state levels of RNAs in the indicated mutant strains. Same membrane has been probed with prA as a ladder, and sizes of mRNA1 products, such as B5 (77 nt), B4 (71 nt) and B1 (47 nt) are indicated. Only the dom34 lane is shown. See Supplementary Fig. 6a for the sequence probed by prH. The 5S rRNA served as a loading control. Total RNA from WT cells without (CGA)4-mRNA expression served as a control, noted C. A non-specific band is indicated by an asterisk. c Xrn1 treatment in vitro of total RNA from dom34 or dom34/ski2 mutant cells and northern blot using probe prH. The 5.8S rRNA is a positive control of Xrn1 treatment. d PCR products obtained from 3′-RACE (see also Fig. 3c). Prior to PCR, cDNAs were produced from cells expressing (CGA)4-mRNA. Total RNA from cells without (CGA)4-mRNA expression served as a control. e Sequences obtained after 3′-RACE performed in d on ski2/DOM34 total RNA. Sequence distribution is given in percentage. f Primer extension experiments using probe prJ to determine the 5′-end of RNAs. Xrn1-specific arrests are indicated by arrowheads. g Positioning of 3′-ends detected by 3′-RACE on (CGA)4-mRNA from ski2/DOM34 cells (magenta arrowhead). Arrowhead sizes are proportional to the relative number of sequences obtained. Three cleavage clusters, C1, C2 and C3 were defined (see Supplementary Fig. 6d). Xrn1 arrests deduced from f are indicated by black arrowhead with sizes proportional to the intensity of reverse stops observed in f. h Schematic view of the ribosome positioning on (CGA)4-mRNA deduced from Xrn1 arrests combined with the positioning of endonucleolytic cleavages provided by 3′-RACE. Source data are provided as a Source Data file.