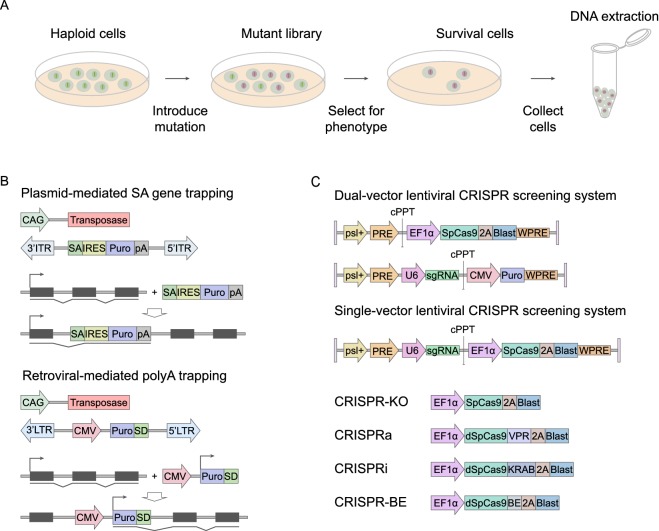
Figure 3.
Applying haploid stem cells for functional genomics. (A) Forward genetic screens are powerful tools for the discovery and functional annotation of genetic elements. Three major steps are: mutagenesis to generate high-throughput mutant libraries, selection for the phenotype of interest, and mapping of mutations. (B) Two types of delivery systems used in gene trapping, the plasmid system and the retroviral system. (Top) Schematic diagram of splice acceptor (SA) gene trap, in which transposon elements were integrated in a plasmid vector. Splicing to upstream exons results in gene trap fusion transcript from which puromycin (puro) is transcribed by an endogenous promoter. CAG, a constitutive promoter; IRES, internal ribosome entry site; pA, poly A; ITR, inverted terminal repeat. (Bottom) Schematic diagram of ployA-trap, in which transposon elements were integrated in a retroviral vector. Insertion of a constitutive promoter-driven marker gene into introns results in a gene trap fusion transcript from which puromycin (puro) is terminated by an endogenous polyadenylation site. CMV, a constitutive promoter; SD, splice donor. LTR, long terminal repeat. (C) Schematic diagram of lentiviral expression vector for SpCas9 and sgRNA in a dual-vector form and single-vector form. psl+, Psi packaging singal; PRE, Rev response element; cPPT, central polypurine tract; EF1α, elongation factor 1α promoter; CMV, immediate-early cytomegalovirus enhancer-promoter; U6, RNA polymerase III U6 promoter; 2A, 2A self-cleavage peptide; Blast, blasticidin selection marker; Puro, puromycin selection marker; WPRE, post-transcriptional regulatory element