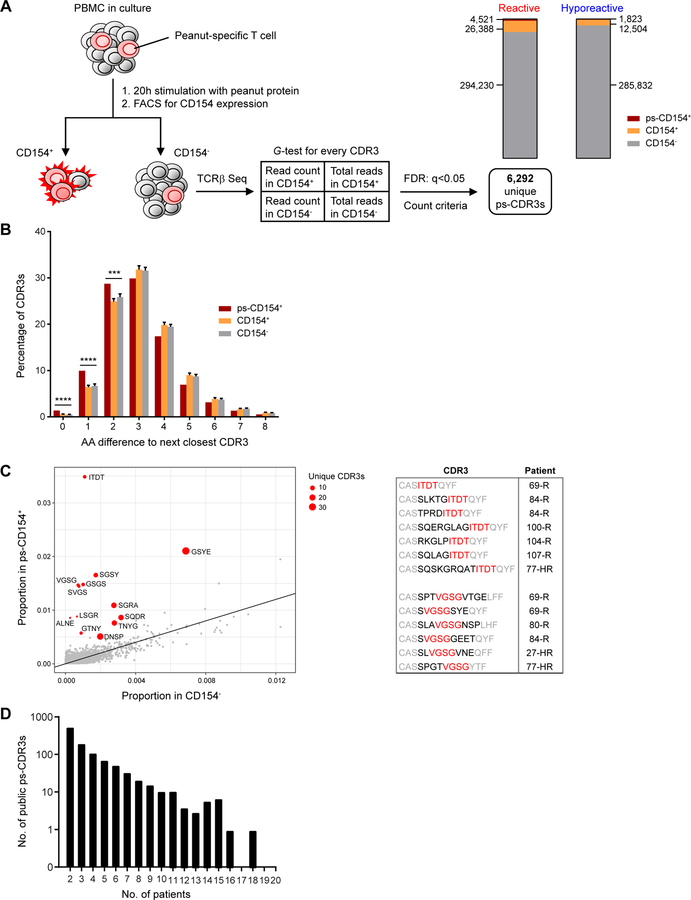
Fig. 1: Enrichment and selection of putatively peanut-specific TCRβ CDR3 sequences.
(A) Procedure for selection of significantly enriched, putatively peanut-specific CDR3 sequences (see Methods and Results). Stacked bar graphs show the proportions and numbers of CDR3s in the CD154−, CD154+, and putatively peanut-specific CD154+ (ps-CD154+) compartments in reactive and hyporeactive patients. (B) Minimum Hamming distance of ps-CDR3s (ps-CD154+), compared with equal-sized randomly sampled control pools of CDR3s from all CD154+ T cells or all CD154− T cells. s.d. of 100 repeat random samples of control CDR3s is shown on bars (**** P < 0.0001, *** P < 0.001, Fisher’s exact test). (C) ps-CDR3s were enriched for a subset of 4-mer amino acid motifs, as compared to CDR3s from CD154− T cells. Shown in red are motifs that were found in at least three unique ps-CDR3s, derived from at least three patients, and met a G-test and FDR cutoff of q < 0.05. The table shows the position of two of the 4-mers (in red) within the ps-CDR3s, and the patients from whom the ps-CDR3s were derived (R = reactive, HR = hyporeactive). Residues with high probability of contact with antigenic peptide are in red and black, those with low probability are in grey. (D) Distribution of the public ps-CDR3s over the patients. Shown is the number of public ps-CDR3s present in a given number of patients.