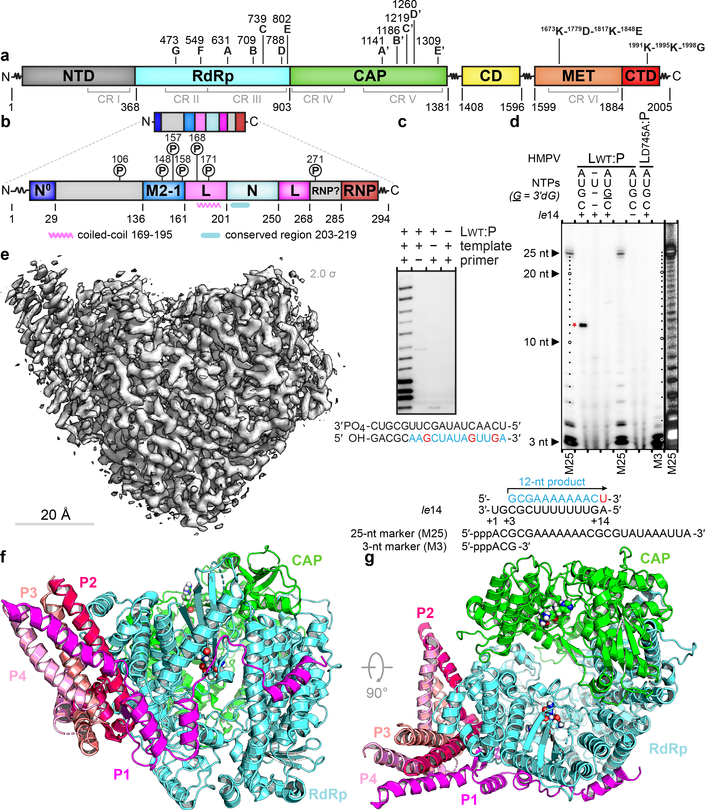
Figure 1. Overall structure of the HMPV L:P complex.
Domain organization of L (a) and P (b) outlining conserved regions and motifs. Regions of HMPV-P predicted to interact with N0, M2–1, L and RNP, and phosphorylation sites are indicated11. (c) Primer elongation assay. Sequences of the 18 nt RNA template and for the 5 nt primer are shown with nascent RNA in blue and radiolabel incorporation sites in red. This experiment was performed a total of four times with two different buffer conditions. (d) RdRp activity assay using the “le14” RNA template. Sequences for the 5’-triphosphorylated 25 nt and 3 nt markers and the 12 nt product are indicated. The 3’dGTP chain terminator is labeled “G”. Radiolabeled UMP is in red. Data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1. (e) Overview of the HMPV-L:P cryo-EM 3D reconstruction (f) Overview of the L:P atomic model. RdRp: cyan, Capping domain: green, phosphoprotein tetramer subunits P1 magenta, P2 hot pink, P3 salmon and P4 pink. Atoms from the “GDNQ” motif in the RdRp and from the “HR” motif in the capping domains are shown as colored spheres (g) Rotated view of the L:P atomic model.