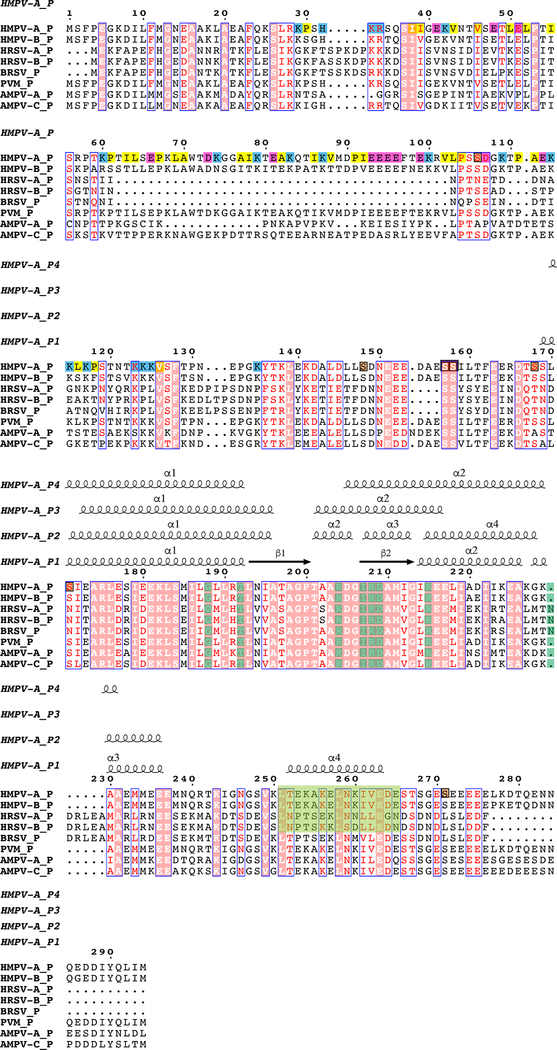
Extended Data Fig. 9 |. Structure-based sequence alignment of the phosphoprotein from HMPV (labeled HMPV-A, strain CAN97-83) and other known pneumoviral P proteins:
HMPV-B, human metapneumovirus subgroup B; HRSV-A and B, human respiratory syncytial virus subgroup A and B respectively; BRSV, bovine respiratory syncytial virus; PVM, pneumonia virus of mice; AMPV-A and AMPV-C, avian metapneumovirus subgroup A and C respectively. Sequences accession codes for the alignment HMPV-A: AAQ67693.1 (used in this work), HMPV-B: AAQ67684.1, HRSV-A: AAX23990.1, HRSV-B: AAR14262.1, BRSV: AAL49395.1, PVM: AAW79177.1, AMPV-A: AAT68644.1 and AMPV-C: AAT86110.1.
The secondary structure of HMPV_P subunit P1 (this work) is displayed above the alignment. Phosphorylation sites are highlighted in brown. Positively-charged residues of HMPV_P are shaded in blue, negatively charged residues in magenta and hydrophobic residues 29 to 135 in yellow. The conserved region containing hydrophobic residues critical for L:P interactions are highlighted in green. Structural alignment of P from HMPV and RSV16 showed similar overall tetramer organization. However, differences are observed in subunit P1 with an r.m.s.d. of 2.24 Å over 82 residues. Although P is in general more mobile with weaker densities and higher B factors compared to L, the region following the beta-hairpin (residues 175–215 in HMPV) does adopt a slightly different conformation compared to RSV P1. Subunit P3 has an r.m.s.d. of 1.94 Å over 45 residues due to a slightly tilted C-terminal helix compared to RSV. Subunit P2 is most similar with an r.m.s.d. of 0.92 Å over 56 residues. Subunit P4 has an r.m.s.d. of 1.33 Å over 47 residues. The eight residues of HRSV-P, that are crucial for interacting with HRSV-L and whose substitutions impair viral replication, are shaded in dark green (data from reference 16). With the exception of Asn189 (HRSV-P) where a deletion is present in HMPV-P, these residues are conserved in HMPV-P and other known pneumoviral P proteins.