Abstract
Background: Numerous deregulated long non-coding RNAs (lncRNAs) accompany the initiation and progression of carcinogenesis. The present study aimed to explore the prognostic significance of lncRNA-SNHG3 on intrahepatic cholangiocarcinoma (ICC) patients. Methods: LncRNA microarray assays were used to evaluate lncRNA expression profiling in three pairs of ICC tissues and adjacent non-tumorous tissues. RT-qPCR was performed to further validate the accuracy of the microarray results. Results: lncRNA microarray and RT-qPCR assays revealed that SNHG3 expression levels were significantly increased in ICC tissues compared to the adjacent non-tumor tissues. A high SNHG3 expression level was significantly correlated with shorter OS in ICC patients. A multivariate regression analysis discovered that SNHG3 could serve as an independent prognostic factor for predicting the OS of ICC patients. Conclusion: We found SNHG3 to be an independent risk factor for predicting the prognosis of ICC. SNHG3 shows a strong promise in the development of novel therapeutic targets for the treatment of ICC.
Keywords: Intrahepatic cholangiocarcinoma, long non-coding RNA, SNHG3, prognosis
Introduction
Intrahepatic cholangiocarcinoma (ICC), frequently located in the biliary tree with epithelial cell malignant growth, is one of the most fatal and the second most common primary hepatic malignant tumor worldwide [1,2]. Patients with ICC trend to be diagnosed at an advanced stage, and the five-year survival rate of ICC is only 10% [3]. At present, surgical resection is the most effective therapeutic method to improve the survival rate, while surgical operations are only being performed on a minority of ICC patients who present at an early stage [4]. Therefore, exploring novel and specific prognostic indicators is very meaningful for improving the survival outcome in ICC patients.
Current biomarkers, such as carbohydrate antigen 19-9 (CA19-9), CA125, carcinoembryonic antigen (CEA), cytokeratin 7 (CK7) and CK19 as the classic biomarkers, are commonly used in the management of patients with ICC [5-8]. However, the sensitivity and specificity of these markers have been questioned. With the development of microarray and sequencing technology, only 2% of the genome constitutes protein-coding genes in mammals, so numerous non-coding RNAs seemingly are “junk genes” [9]. In fact, non-coding RNAs, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs, have attracted much attention due to their large mass and numerous biological functions, including the regulation of transcription and translation, and their function as competing endogenous RNAs and regulators of carcinogenesis [10,11]. Recently, non-coding RNAs are thought to be independent prognostic biomarkers in various cancer types [12-14]. These studies suggest that non-coding RNAs’ potential as prognostic biomarkers may facilitate the translation of basic research into clinical practice.
lncRNAs are characterized by their length of more than 200 bp and their non-protein encoding transcript and have been validated frequently by their dysregulated expression in a variety of tumors, including CCA [15]. Intriguingly, many lncRNAs have been reported as potential prognostic factors for many human cancers, such as hepatocellular carcinoma [16], non-small cell lung cancer [17], and breast cancer [18]. To our knowledge to date, several lncRNAs, including CCAT, TUG1, and CRNDE, may serve as potential molecular biomarkers for predicting the prognosis of ICC [4,19,20]. In the present study, we examined the lncRNA expression profiles in ICC and adjacent non-tumorous tissues to investigate their potential use as prognostic markers of ICC. We found that lncRNA-SNHG3 was elevated in tumor tissues from ICC patients. More importantly, lncRNA-SNHG3 could function as an independent prognostic biomarker of ICC.
Material and methods
Patients and specimens
A total of fifty-two ICC patients were enrolled in our study from January 2009 to June 2012. Tissue-based specimens were collected from fifty-two ICC patients who had undergone surgery. None of the patients enrolled received any radiotherapy or chemotherapy before their operations. All the tumors from the ICC patients were clinically and histologically diagnosed with ICC. All specimens were immediately preserved in liquid nitrogen for experimental measurements. Signed informed consent forms were obtained from all ICC patients. Our study was approved by the Ethics Committee of the Second Affiliated Hospital of Kunming Medical University (Kunming, China) according to the Helsinki Declaration.
LncRNAs microarray
Differentially expressed lncRNAs in three pairs of ICC tissues and adjacent non-tumorous tissues were analyzed using the Agilent Human lncRNA Array V.2.0 platform (Agilent Technologies, Santa Clara, CA, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using RNAiso (Takara, Dalian, China). An RT Kit (Toyobo Co. Ltd, Osaka, Japan) and KAPA SYBR Green FAST qPCR Kit (KAPA, Wilmington, MA, US) were used to measure the expression levels of lncRNAs using an Applied Biosystems 7300 Real-Time PCR System (Thermo Fisher Scientific, Inc.) and normalized to the internal control U6. The lncRNA expression levels were calculated using the 2-ΔΔCq method [21].
Statistical analysis
Data were presented as the mean ± the standard error of the mean. The statistical analysis was performed using SPSS software version 19.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism Version 7.0 (GraphPad Software, Inc., La Jolla, CA, USA). A Student’s t-test or a Kruskal-Wallis test was used to analyze the two-group differences. Pearson χ2 tests were used to evaluate differences between the clinical characteristics and lncRNA-SNHG3 expression levels in the ICC tissues. Overall survival (OA) was calculated using the Kaplan-Meier method with the log-rank test applied for comparison. Univariate and multivariate regression analyses were performed to evaluate the correlation between the lncRNA-SNHG3 expression and OS using the Cox proportional hazard model. P < 0.05 was considered to indicate a statistically significant difference.
Results
Differentially expressed lncRNAs in human ICC tissues
Differentially expressed lncRNAs in three pairs of human ICC and adjacent non-tumorous tissues were measured using a lncRNA microarray analysis. The results revealed that the 30 mostly significantly dysregulated lncRNAs were picked out, according to the Log2|FC| ≥ 2.0, P < 0.001 and FDR < 0.001. Among these lncRNAs, 13 and 17 lncRNAs were down-regulated and up-regulated, respectively, in human ICC tissues compared with the adjacent non-tumorous tissues (Figure 1A). To further validate the accuracy of the microarray results, the top 5 differentially expressed lncRNAs, including PCAT1, SNHG16, SNHG3, LASP1-AS, and LINC-USP16, were analyzed using RT-qPCR assays. PCAT1, SNHG16, SNHG3, LASP1-AS expression levels were significantly increased, but LINC-USP16 expression levels were markedly decreased in fifty-two ICC tissues as compared to adjacent non-tumor tissues (Figure 1B). These results were consistent with the microarray assay. We also found the fold change (FC) of SNHG3 at the maximum value in the top 5 differentially expressed lncRNAs using RT-qPCR assays. Therefore, we focused on SNHG3 in the subsequent experiments.
Figure 1.
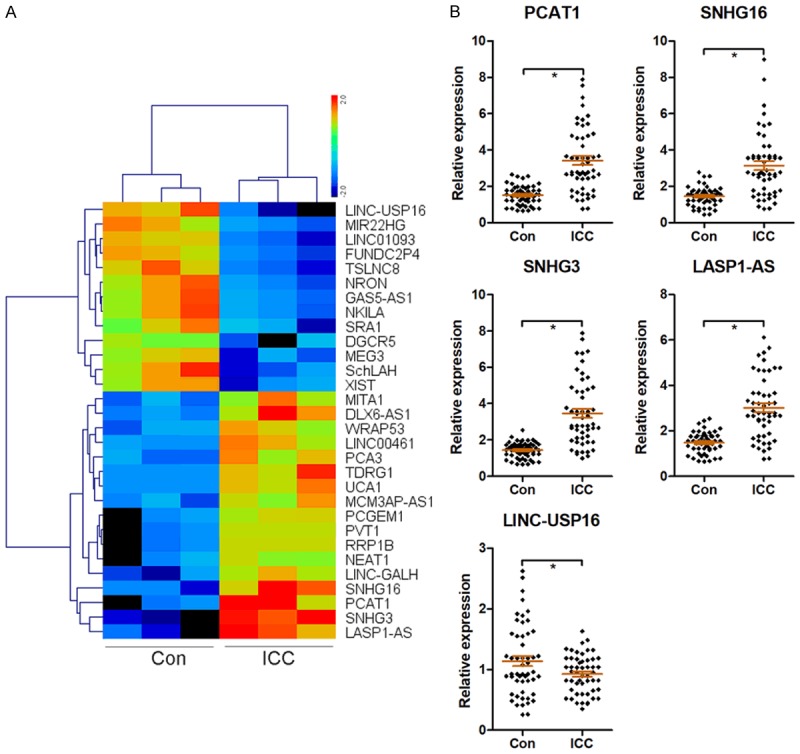
Differentially expressed lncRNAs in human ICC tissues. LncRNA expression profiling in three pairs of human ICC and adjacent non-tumorous tissues was analyzed using an lncRNA microarray, and a heatmap of 30 differentially expressed lncRNAs was presented based on Log2|FC| ≥ 2.0, P < 0.001 and FDR < 0.001 (A). The top 5 differentially expressed lncRNAs, including PCAT1, SNHG16, SNHG3, LASP1-AS, and LINC-USP16, were analyzed using RT-qPCR assays (B). *P < 0.05.
SNHG3 is an independent prognostic biomarker of ICC patients
The relationship between SNHG3 expression and the clinicopathological characteristics of ICC patients is shown in Table 1. Pearson χ2 tests indicated that the expression levels of SNHG3 had a significant correlation with age, gender, serum CA19-9 levels, tumor size, and histological differentiation. However, the up-regulation of the SNHG3 expression level is significantly correlated with poor TNM stage (P = 0.006), lymph node metastasis (P = 0.008), and distant metastasis (P = 0.010) in ICC patients. In addition, the Kaplan-Meier method and the log-rank test showed that age, gender, serum CA19-9 levels, tumor size, and histological differentiation could not be used as indicators for predicting OS of ICC patients (Figure 2). Poor TNM stage (P = 0.0086), lymph node metastasis (P = 0.0009), distant metastasis (P = 0.0215) and high SNHG3 expression levels (P = 0.0094) were significantly correlated with short OS in ICC patients (Figure 2). Univariate and multivariate regression analyses showed that TNM stage, lymph node metastasis, distant metastasis, and SNHG3 could serve as independent prognostic factors for predicting the OS of ICC patients (Table 2).
Table 1.
Correlations between the lncRNA-SNHG3 expression levels and the clinicopathological variables of ICC patients
| Variables | n | LncRNA-SNHG3 expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low (n = 25) | High (n = 27) | |||
| Age (years) | 0.554 | |||
| ≤ 60 | 29 | 15 | 14 | |
| > 60 | 23 | 10 | 13 | |
| Gender | 0.586 | |||
| M | 27 | 12 | 15 | |
| F | 25 | 13 | 12 | |
| CA19-9 (U/mL) | 0.618 | |||
| ≤ 40 | 19 | 10 | 9 | |
| > 40 | 33 | 15 | 18 | |
| Tumor size (cm) | 0.376 | |||
| ≤ 3 | 30 | 16 | 14 | |
| > 3 | 22 | 9 | 13 | |
| TNM stage | 0.006 | |||
| I/II | 23 | 16 | 7 | |
| III/IV | 29 | 9 | 20 | |
| Histological differentiation | 0.262 | |||
| Well/Moderate | 27 | 15 | 12 | |
| Poor | 25 | 10 | 15 | |
| Lymph node metastasis | 0.008 | |||
| No | 32 | 20 | 12 | |
| Yes | 20 | 5 | 15 | |
| Distant metastasis | 0.010 | |||
| No | 37 | 22 | 15 | |
| Yes | 15 | 3 | 12 | |
Figure 2.
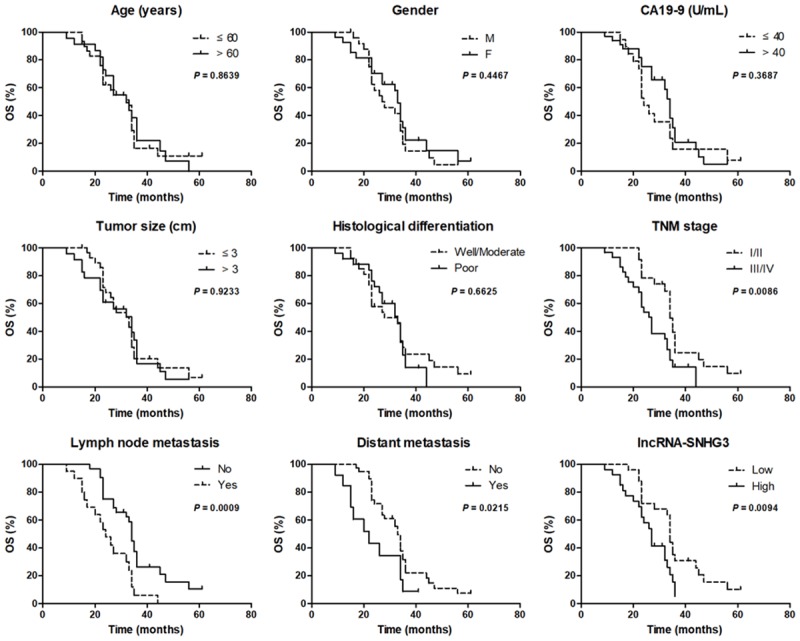
SNHG3 was an independent prognostic biomarker for ICC patients. A Kaplan-Meier survival curve analysis and a log-rank test were applied to evaluate whether age, gender, CA19-19, tumor size, histological differentiation, TNM stage, lymph node metastasis, distant metastasis and lncRNA-SNHG3 expression levels were correlated with the OS of ICC patients.
Table 2.
Univariate and multivariate regression analysis of ICC patients for overall survival
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (> 60 vs ≤ 60) | 1.23 (0.67-2.90) | 0.755 | ||
| Gender (M vs F) | 1.31 (0.75-3.16) | 0.562 | ||
| CA19-9 (> 40 vs ≤ 40) | 1.53 (0.86-3.79) | 0.349 | ||
| Tumor size (> 3 vs ≤ 3) | 1.27 (0.70-3.05) | 0.647 | ||
| Histological differentiation (Poor vs Well/Moderate) | 1.48 (0.81-3.42) | 0.455 | ||
| TNM stage (III/IV vs I/II) | 3.72 (1.84-8.93) | 0.003 | 2.95 (1.42-7.37) | 0.011 |
| Lymph nodes metastasis (Yes vs No) | 3.44 (1.66-7.94) | 0.007 | 2.48 (1.11-5.73) | 0.019 |
| Distant metastasis (Yes vs No) | 2.38 (1.04-5.56) | 0.022 | 2.09 (0.96-4.87) | 0.041 |
| LncRNA-SNHG3 (high vs low) | 3.25 (1.54-7.51) | 0.009 | 2.82 (1.36-7.03) | 0.015 |
Discussion
ICC is one of the most infamous malignant tumors in humans and is usually diagnosed at the advanced stages or through distant metastasis, which leads to a poor prognosis [22]. More reliable and effective prognostic markers may provide new ideas for improving the clinical outcomes in ICC patients. Recent studies have demonstrated that numerous lncRNAs are deregulated in ICC [23,24]. For example, Wang et al. revealed that 2773 lncRNAs and 2392 lncRNAs were significantly up-regulated and down-regulated, respectively, in ICC tissues compared with the noncancerous tissues [23]. Lv et al. performed an lncRNA microarray assays in four pairs of ICC tissues and normal tissues, and found that 2716 lncRNAs are markedly differentially expressed in ICC tissues compared with normal tissues [24]. Moreover, several lncRNAs, such as CCAT1, CRNDE, and CCAT2, promote the initiation and progression of ICC, and the up-regulation of these lncRNAs are associated with poor OS in ICC patients [20,25,26].
In our study, SNHG3, as a prominent carcinogenic lncRNA, was significantly increased in ICC tissues compared with adjacent non-tumor tissues, which was validated by a lncRNA microarray and by RT-qPCR assays. Importantly, SNHG3 was identified as an independent prognostic indicator for OS in ICC patients. SNHG3 as an oncogene has been reported in several cancer types, such as osteosarcoma, lung adenocarcinoma, hepatocellular carcinoma, ovarian cancer, and colorectal cancer [27-31]. In two independent cohort studies, patients with high SNHG3 expressions had poorer OS than those with a low expression of SNHG3 in ovarian cancer and osteosarcoma [27,30]. The TCGA database also shows that high SNHG3 expression is correlated with poor OS in colorectal cancer and lung adenocarcinoma [28,31]. We also found that ICC patients with high SNHG3 expression exhibited a significant shorter OS than patients with low SNHG3 expression. Moreover, the up-regulation of SNHG3 expression was significantly positively related with poor TNM stage, lymph node metastasis and distant metastasis in ICC patients. Multivariate regression analysis validated that poor TNM stage, lymph node metastasis and distant metastasis could also serve as independent prognostic factors of ICC.
However, there are some limitations to our study. First, the relatively low number of cases may impact the credibility of the conclusions in our study. The functional aspect of lncRNA usually competes with endogenous RNA to sponge miRNA and its downstream target gene [32]. However, we did not investigate the roles of SNHG3 on ICC cells growth in vitro and in vivo.
In summary, we found SNHG3 to be an independent risk factor for predicting the prognosis of ICC. SNHG3 looks promising as a means of developing a novel therapeutic target for the treatment of ICC.
Acknowledgements
This research was supported by the Project for Research Institutions of Yunnan Provincial Health Bureau (grant. no. 2016ns249) and the Scientific Research Foundation of Yunnan Provincial Education Bureau of China (grant. no. 2015C042Y).
Disclosure of conflict of interest
None.
References
- 1.Wang WT, Ye H, Wei PP, Han BW, He B, Chen ZH, Chen YQ. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9:117. doi: 10.1186/s13045-016-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skipworth JR, Olde Damink SW, Imber C, Bridgewater J, Pereira SP, Malago M. Review article: surgical, neo-adjuvant and adjuvant management strategies in biliary tract cancer. Aliment Pharmacol Ther. 2011;34:1063–1078. doi: 10.1111/j.1365-2036.2011.04851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Y, Leng K, Li Z, Zhang F, Zhong X, Kang P, Jiang X, Cui Y. The prognostic potential and carcinogenesis of long non-coding RNA TUG1 in human cholangiocarcinoma. Oncotarget. 2017;8:65823–65835. doi: 10.18632/oncotarget.19502. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.He C, Zhang Y, Song Y, Wang J, Xing K, Lin X, Li S. Preoperative CEA levels are supplementary to CA19-9 levels in predicting prognosis in patients with resectable intrahepatic cholangiocarcinoma. J Cancer. 2018;9:3117–3128. doi: 10.7150/jca.25339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loosen SH, Roderburg C, Kauertz KL, Koch A, Vucur M, Schneider AT, Binnebosel M, Ulmer TF, Lurje G, Schoening W, Tacke F, Trautwein C, Longerich T, Dejong CH, Neumann UP, Luedde T. CEA but not CA19-9 is an independent prognostic factor in patients undergoing resection of cholangiocarcinoma. Sci Rep. 2017;7:16975. doi: 10.1038/s41598-017-17175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CY, Shiesh SC, Tsao HC, Lin XZ. The assessment of biliary CA 125, CA 19-9 and CEA in diagnosing cholangiocarcinoma--the influence of sampling time and hepatolithiasis. Hepatogastroenterology. 2002;49:616–620. [PubMed] [Google Scholar]
- 8.Liu LZ, Yang LX, Zheng BH, Dong PP, Liu XY, Wang ZC, Zhou J, Fan J, Wang XY, Gao Q. CK7/CK19 index: a potential prognostic factor for postoperative intrahepatic cholangiocarcinoma patients. J Surg Oncol. 2018;117:1531–1539. doi: 10.1002/jso.25027. [DOI] [PubMed] [Google Scholar]
- 9.Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, Calin GA. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–5011. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012;11:189–200. doi: 10.1016/S1474-4422(11)70286-1. [DOI] [PubMed] [Google Scholar]
- 11.Xie H, Ma B, Gao Q, Zhan H, Liu Y, Chen Z, Ye S, Li J, Yao L, Huang W. Long non-coding RNA CRNDE in cancer prognosis: review and meta-analysis. Clin Chim Acta. 2018;485:262–271. doi: 10.1016/j.cca.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Miao R, Ge C, Zhang X, He Y, Ma X, Xiang X, Gu J, Fu Y, Qu K, Liu C, Wu Q, Lin T. Combined eight-long noncoding RNA signature: a new risk score predicting prognosis in elderly non-small cell lung cancer patients. Aging (Albany NY) 2019;11:467–479. doi: 10.18632/aging.101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang D, Peng Y, Ma K, Deng X, Tang L, Jing D, Shao Z. MiR-20a, a novel promising biomarker to predict prognosis in human cancer: a meta-analysis. BMC Cancer. 2018;18:1189. doi: 10.1186/s12885-018-4907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Yao Y, Jiang X, Zhong X, Wang Z, Li C, Kang P, Leng K, Ji D, Li Z, Huang L, Qin W, Cui Y. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J Exp Clin Cancer Res. 2018;37:81. doi: 10.1186/s13046-018-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Z, Zhou F, Yang Y, Li L, Lei Y, Lin X, Li H, Pan X, Chen J, Wang G, Liu H, Jiang J, Wu B. Lnc-PCDH9-13:1 is a hypersensitive and specific biomarker for early hepatocellular carcinoma. EBioMedicine. 2018;33:57–67. doi: 10.1016/j.ebiom.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo CL, Xu ZG, Chen H, Ji J, Wang YH, Hu W, Wang K, Zhang WW, Yuan CH, Wang FB. LncRNAs and EGFRvIII sequestered in TEPs enable blood-based NSCLC diagnosis. Cancer Manag Res. 2018;10:1449–1459. doi: 10.2147/CMAR.S164227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amorim M, Salta S, Henrique R, Jeronimo C. Decoding the usefulness of non-coding RNAs as breast cancer markers. J Transl Med. 2016;14:265. doi: 10.1186/s12967-016-1025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai K, Quan J, Yan F, Jin X, Pan X, Song X, Zhang S, Ren Q, Liu J, Liu X. lncRNAs as potential molecular biomarkers in the clinicopathology and prognosis of cholangiocarcinoma: a systematic review and meta-analysis. Onco Targets Ther. 2019;12:1905–1915. doi: 10.2147/OTT.S188134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia XL, Xue D, Xiang TH, Xu HY, Song DK, Cheng PG, Wang JQ. Overexpression of long non-coding RNA CRNDE facilitates epithelial-mesenchymal transition and correlates with poor prognosis in intrahepatic cholangiocarcinoma. Oncol Lett. 2018;15:4105–4112. doi: 10.3892/ol.2018.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Peng T, Li Z, Li D, Wang S. MACC1 promotes angiogenesis in cholangiocarcinoma by upregulating VEGFA. Onco Targets Ther. 2019;12:1893–1903. doi: 10.2147/OTT.S197319. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Wang J, Xie H, Ling Q, Lu D, Lv Z, Zhuang R, Liu Z, Wei X, Zhou L, Xu X, Zheng S. Coding-noncoding gene expression in intrahepatic cholangiocarcinoma. Transl Res. 2016;168:107–121. doi: 10.1016/j.trsl.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Lv L, Wei M, Lin P, Chen Z, Gong P, Quan Z, Tang Z. Integrated mRNA and lncRNA expression profiling for exploring metastatic biomarkers of human intrahepatic cholangiocarcinoma. Am J Cancer Res. 2017;7:688–699. [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Xiao J, Chai Y, Du YY, Liu Z, Huang K, Zhou X, Zhou W. LncRNA-CCAT1 promotes migration, invasion, and EMT in intrahepatic cholangiocarcinoma through suppressing miR-152. Dig Dis Sci. 2017;62:3050–3058. doi: 10.1007/s10620-017-4759-8. [DOI] [PubMed] [Google Scholar]
- 26.Bai JG, Tang RF, Shang JF, Qi S, Yu GD, Sun C. Upregulation of long noncoding RNA CCAT2 indicates a poor prognosis and promotes proliferation and metastasis in intrahepatic cholangiocarcinoma. Mol Med Rep. 2018;17:5328–5335. doi: 10.3892/mmr.2018.8518. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Wu Z, Zhang Y. LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma. Int J Immunopathol Pharmacol. 2019;33:2058738418820743. doi: 10.1177/2058738418820743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Ni J, He X. Upregulation of the long noncoding RNA SNHG3 promotes lung adenocarcinoma proliferation. Dis Markers. 2018;2018:5736716. doi: 10.1155/2018/5736716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang PF, Wang F, Wu J, Wu Y, Huang W, Liu D, Huang XY, Zhang XM, Ke AW. LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J Cell Physiol. 2019;234:2788–2794. doi: 10.1002/jcp.27095. [DOI] [PubMed] [Google Scholar]
- 30.Hong L, Chen W, Wu D, Wang Y. Upregulation of SNHG3 expression associated with poor prognosis and enhances malignant progression of ovarian cancer. Cancer Biomark. 2018;22:367–374. doi: 10.3233/CBM-170710. [DOI] [PubMed] [Google Scholar]
- 31.Huang W, Tian Y, Dong S, Cha Y, Li J, Guo X, Yuan X. The long non-coding RNA SNHG3 functions as a competing endogenous RNA to promote malignant development of colorectal cancer. Oncol Rep. 2017;38:1402–1410. doi: 10.3892/or.2017.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]


