Figure 1.
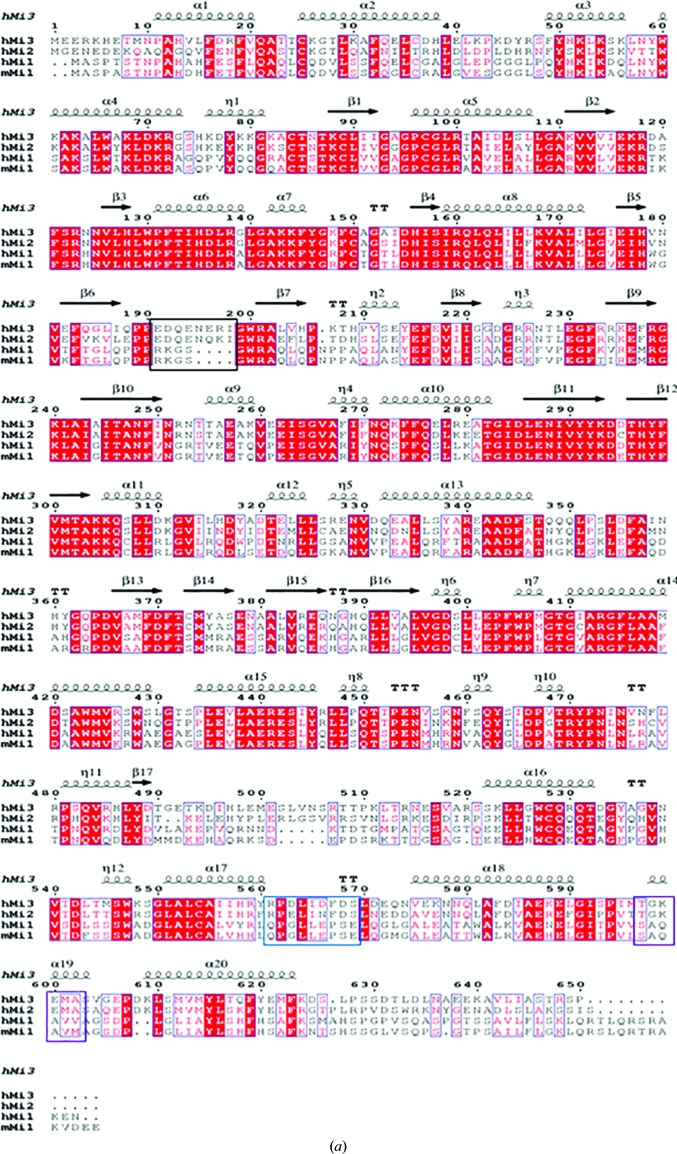
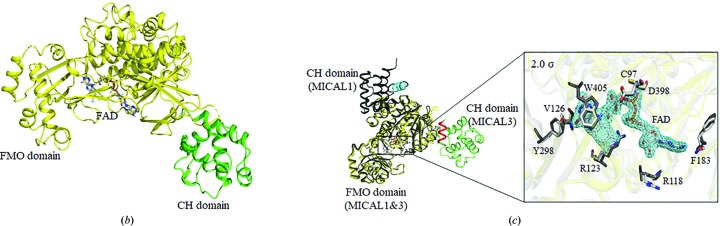
Overall structure and characterization of human MICAL3FMOCH. (a) Sequence alignment of human MICAL3, human MICAL2, human MICAL1 and mouse MICAL1. Strictly conserved residues are boxed in red, while similar residues are shown as red letters. The sequence-alignment tools used were ClustalW and ESPript. 310-Helices are represented by η, strict β-turns are represented by TT and strict α-turns by TTT. (b) The crystal structure of human MICAL3. The N- and C-termini are labeled N and C, respectively. The FMO domain is shown in yellow and the CH domain is in green. (c) The FAD-binding site. FAD is shown as a stick model and the 2F o − F c map for the FAD molecule is contoured at 2σ. The distances between residues and FAD were calculated using PISA. The stick model of human MICAL3 is shown in yellow, white, red and blue, whereas the ribbon model is shown in yellow; the stick and ribbon models of mouse MICAL1 are shown in black.