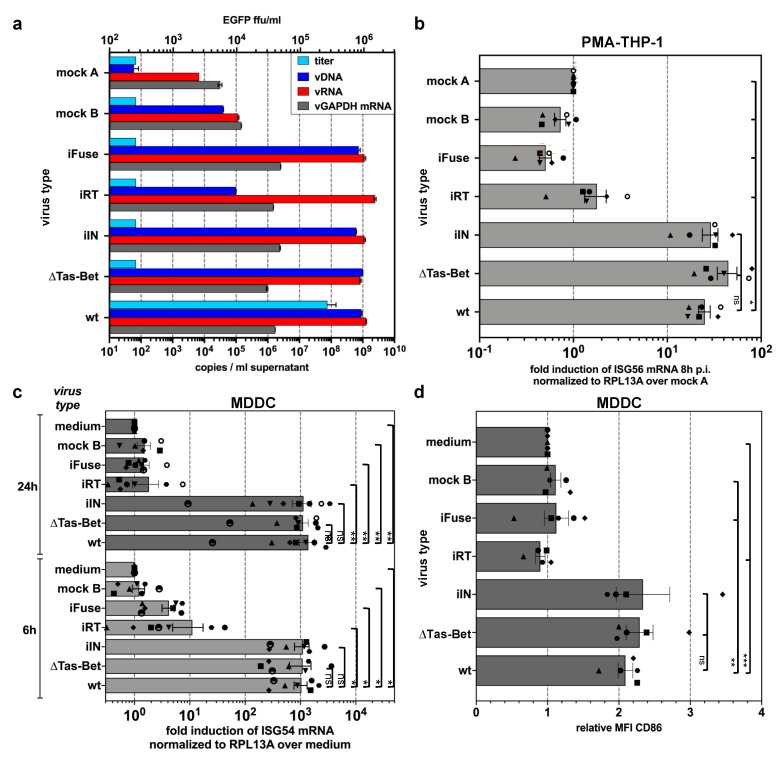
Figure 3.
Differential ISG induction profiles of PFV mutants (described in detail in Figure A1 and Material and Methods) varying in their protein and nucleic acid composition. (a) Particle-associated nucleic acids extracted from viral particles pelleted by ultracentrifugation of virus supernatants used in (b–d) were analyzed by qPCR to quantify the particle-associated viral (vDNA: viral DNA; vRNA: viral RNA) and cellular (vGAPDH mRNA) nucleic acid composition. Mean copy numbers ± SDs per mL supernatant determined from duplicates are shown. (b,c) ISG induction profile of PMA-differentiated THP-1 wild type cells (b) or MDDCs (c) incubated with identical amounts of wild type PFV-RCP (MOI 0.1 THP-1; MOI 0.25 MDDCs) supernatants, variants thereof, and pUC19 (mock A) and ∆GPE (mock B) mock supernatants, or medium, as indicated, for 8 h and 24 h. Means ± SEMs, plus individual data points, of ISG56 (n = 6) or ISG54 (n = 5–7) mRNA induction normalized to RPL13A mRNA relative to mock A or medium treatment are shown. One-way ANOVA with Tukey’s multiple-comparisons test (b) or mixed-effects analysis with Tukey’s multiple-comparisons test (c) was used to assess significance. (d) CD86 cell surface expression profile of MDDCs 24 h post exposure to wild type PFV-RCP (MOI 0.25) supernatants and variants thereof as indicated. Means ± SEMs (n = 5), plus individual data points, relative to medium treatment are shown. Mixed-effects analysis with Tukey’s multiple-comparisons test was used to assess significance. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; ns: not significant (p ≥ 0.05).