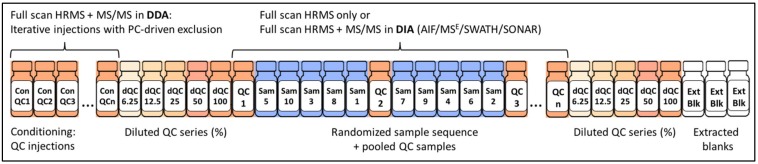
Figure 2.
Setting up the data acquisition worklist to facilitate metabolite quantification and identification. Prior to batch run, the instrument should be conditioned (or “passivated”) using the pooled quality control (QC) of biological samples. During the conditioning, high-quality MS/MS data can be acquired in a data-dependent acquisition (DDA) mode by taking advantage of iterative injections through the application of PC-driven exclusion (of ions for which the MS/MS data have already been acquired). In this way, the amount of acquired high-quality MS/MS data will be maximized. The batch run can start (and end) with the analysis of diluted QC series that will serve to remove the features whose response is not linear; however, this removal should be performed carefully by evaluating low abundance features and those with saturation issues. Finally, samples should be run in a randomized fashion (considering the most important confounding factors, such as disease, sex, age, etc., depending on the experiment) with pooled QCs every 4–10 samples (depending on the size of the batch). Extracted blanks can be analyzed after the sample run and used for the removal of background (chemical and informatic) noise. Abbreviations: MS/MS data—fragmentation pattern, HRMS—high-resolution mass spectrometry, DDA—data-dependent acquisition, DIA—data-independent acquisition, AIF—all ion fragmentation (on Agilent or Thermo systems), MSE—all ion fragmentation on Waters systems-, SWATH—sequential window acquisition of all theoretical mass spectra or DIA strategy on Sciex systems, SONAR—scanning quadrupole DIA or DIA strategy on Waters systems.