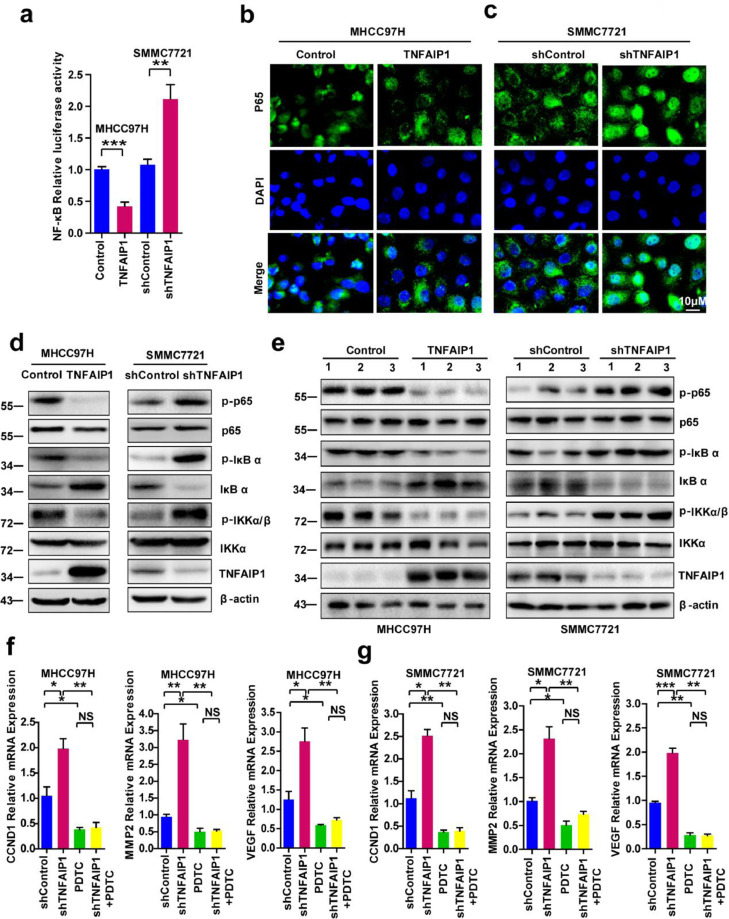
Fig. 5.
TNFAIP1 inhibits the trans-activation of NF-κB. a. The luciferase activity was detected in MHCC97H cells infected with TNFAIP1 or the control lentivirus and in SMMC7721 cells infected with shTNFAIP1 or shControl lentivirus by co-transfected with NF-κB luciferase reporter plasmid. Relative luciferase activities represent mean ± SEM from triplicate independent experiments (**P < 0.01, ***P < 0.001, Student's t-test). b. Immuofluorescence microscopy staining p65 antibody in MHCC97H cells infected with TNFAIP1 or the control lentivirus, the green signal represents p65 staining and blue signal represents nuclear DNA staining by DAPI. Scale bar, 10 μm. c. Immunofluorescence microscopy staining p65 antibody in SMMC7721 cells infected with shTNFAIP1 or shControl lentivirus, the green signal represents p65 staining and blue signal represents nuclear DNA staining by DAPI. Scale bar, 10 μm. d. Western blot analysis of p65, p-p65, IκBα, p-IκBα, IKKα/β and p-IKKα/β in MHCC97H cells infected with TNFAIP1 or the control lentivirus and in SMMC7721 cells infected with shTNFAIP1 or shControl lentivirus. e. Tumors from MHCC97H-TNFAIP1, MHCC97H—Control, SMMC7721-shTNFAIP1 and SMMC7721-shControl groups were used to detect the expression of p65, p-p65, IκBα, p-IκBα, IKKα/β and p-IKKα/β. f. RT-qPCR analysis of the mRNA levels of CCND1, MMP2 and VEGF in MHCC97H cells infected with shControl or shTNFAIP1 lentivirus and treated with PDTC (20 μM) (*P < 0.05, **P<0.01, Student's t-test). g. RT-qPCR analysis of the mRNA levels of CCND1, MMP2 and VEGF in SMMC7721 cells infected with shControl or shTNFAIP1 lentivirus and treated with PDTC (*P<0.05, **P < 0.01, ***P<0.001, Student's t-test). Data are presented as means ± SEM from triplicate independent experiments. P-values were determined by two-tailed Student's t-test (*P < 0.05, **P < 0.01, ***P < 0.001).