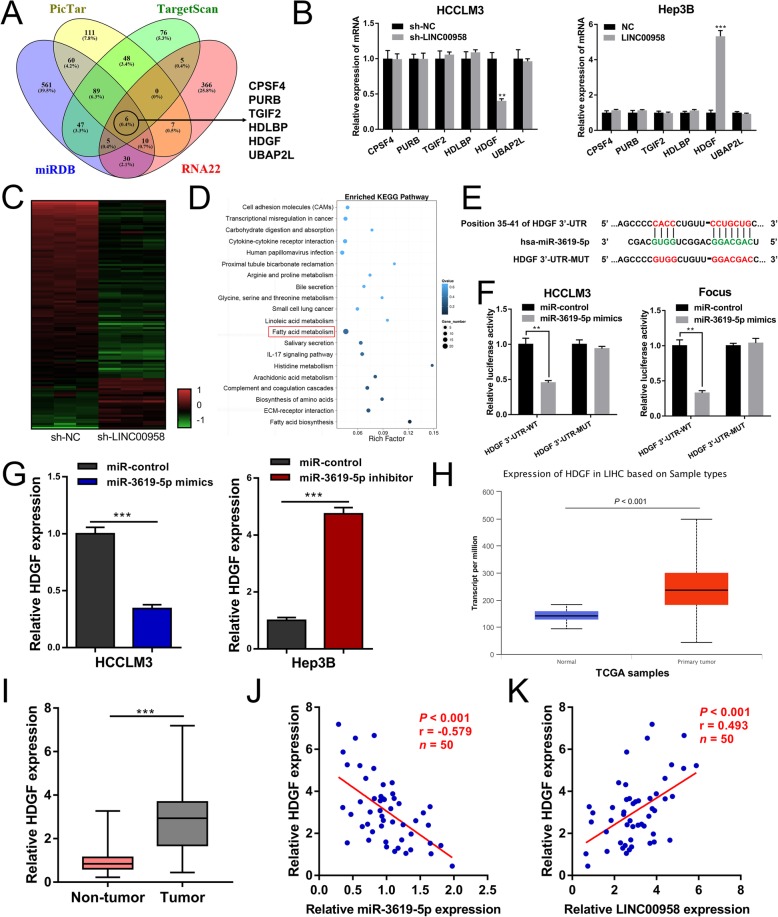
Fig. 4.
HDGF is a direct target gene of miR-3619-5p. a Venn diagram showing six putative miR-3619-5p target genes predicted by four different algorithms (PicTar, TargetScan, miRDB, and RNA22). b RT-qPCR analysis showed that HDGF was downregulated in LINC00958-silenced HCCLM3 cells and upregulated in LINC00958-overexpressed Hep3B cells. c Heat map showing the differentially expressed genes modulated by LINC00958 knockdown. d Enriched KEGG pathway analysis showing the most enriched pathways. e Putative binding sequence of miR-3619-5p in the 3′-UTR of HDGF. f Dual luciferase reporter assay revealed that miR-3619-5p could bind to the 3′-UTR of HDGF. g Expression levels of HDGF were detected by RT-qPCR in miR-3619-5p-overexpressed HCCLM3 cells or miR-3619-5p-silenced Hep3B cells. h TCGA data suggested that the expression level of HDGF was upregulated in liver cancer samples. i RT-qPCR results showing the expression levels of HDGF in 50 paired HCC tissues and non-tumor specimens. j Correlation analysis showing a negative correlation between miR-3619-5p and HDGF expression (P = 0.011). k Expression level of LINC00958 was positively correlated with HDGF expression level (P = 0.002)