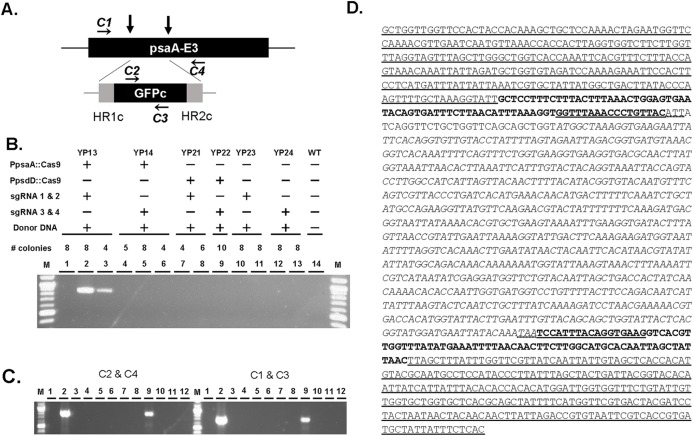
Figure 2. Replacement of the chloroplast genome with donor DNA by Cas9/gRNA.
(A) Schematic view of the psaA-E3 genomic region targeted by two gRNAs (vertical arrows) and the donor DNA composed of codon-optimized GFPc gene that is provided by the Edit Plasmid. The recognition sites of primers used for the amplification of the junction regions are indicated (C1–C4). (B) PCR amplification of the junction region of the replaced DNA. Pooled DNAs extracted from independent colonies was used as templates with primers C2 and C4. Total number of colonies for each Edit Plasmids was 20 for YP13, 17 for YP14, 10 for YP21, 10 for YP22, 16 for YP23 and 24. Template for Lane 14 was untransformed wild-type cells. C2/C4 amplicon was 852 bp long. M: 1 kb plus molecular weight marker (New England Biolabs, Ipswich, MA, USA). (C) De-convolution of junction-PCR positive pools of YP13 transformants. A total of 12 events of the positive pools from Lanes 2 and 3 of (B) were analyzed by two primer sets C2/C4 and C1/C3 to amplify the left and right junction regions, respectively. Events #2 and #9 carried replaced DNA. C1/C3 amplicon was 712 bp long. (D) The sequence obtained from PCR amplification of the replacement DNA locus in Chlamydomonas plastid DNA modified by the Edit Plasmid approach. Underlined sequences: wild-type chloroplast genomic sequence that are not present on the Edit Plasmid. Sequences in bold: homologous regions (HR1c and HR2c) present in the donor DNA on the Edit Plasmid. Sequences in bold underlined: modified gRNA target sites present in the donor DNA. Sequences with double underlines: Silent mutations at the 3′ side of guide RNA sites to preclude re-cleavage by Cas9/sgRNA. (D) The sequence obtained from PCR amplification of the replacement DNA locus in Chlamydomonas plastid DNA modified by the Edit Plasmid approach. Underlined sequences: wild-type chloroplast genomic sequence that are not present on the Edit Plasmid. Sequences in bold: homologous regions (HR1c and HR2c) present in the donor DNA on the Edit Plasmid. Sequences in bold underlined: modified gRNA target sites present in the donor DNA. Sequences with dotted underlines: silent mutations at the 3′ side of guide RNA sites to preclude re-cleavage by Cas9/sgRNA.