Abstract
Background
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm where pathogenesis is based on the oncoprotein termed BCR-ABL1.1 TET2 initiates DNA demethylation and is frequently mutated in hematological malignancies, including CML. The relation between TET2 acquisition and CML transformation and/or imatinib resistance is needed to be investigated.2
Aim
To evaluate Ten Eleven Translocation 2 gene (TET2) single nucleotide polymorphism (SNP) (rs2454206, rs34402524, rs61744960) in chronic myeloid leukemia (CML) in relation to the disease prognostic criteria.
Materials & Method
The study included 84 subjects; 54 CML in chronic phase and 30 healthy subjects as control group matched for age and sex. Routine investigations, including CBC, bone marrow aspiration, biochemical investigations, and molecular study, were performed in CML patients to identify the disease stage. DNA extraction and SNP assay for TET2 gene polymorphism were done using (Thermo-Fisher predesigned SNP, USA) PCR prism 7500.
Results
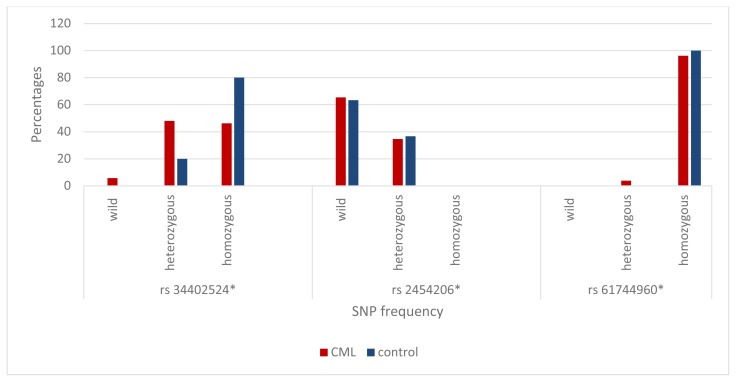
The mean age was 45.98±15.7 yrs in CML patients and 39.3±6.587 yrs in the control group (p>0.05). TET2 SNP rs 34402524 was either heterozygous or homozygous in CML (48%, 46.2% respectively) but was mainly homozygous among control (80%) group (p=0.012). TET2 SNP rs 2454206 wild type within CML was detected in 65.4% of patients and in controls was 63.3% (p=0.046). TET2 SNP rs 61744960 showed a homozygous pattern among all groups (CML and control) (p=0.528). TET2 SNP in CML cases did not alter the prognostic criteria as no statistical significance was noted (p>0.05) yet, it was significantly related to spleen size in rs 34402524 where the homozygous group had larger spleen size and higher BCR-ABL1 levels six months after starting TKIs (p<0.05).
Conclusions/Recommendation
TET2 SNP is common among Egyptian chronic myeloid leukemia. TET2 SNP rs 3442524 was associated with larger spleen size and higher BCR-ABL1 levels after six months of starting TKIs suggesting disease progression.
Keywords: Chronic Myeloid Leukemia, Ten Eleven Translocation 2 Gene
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm. Its pathogenesis is based on the oncoprotein BCR-ABL1 that is the fusion of the Abelson murine leukemia (ABL1) gene on chromosome 9 and the breakpoint cluster region (BCR) gene on chromosome 22. This oncoprotein is an active tyrosine kinase that promotes growth and replication. Several signaling pathways downstream are involved, such as RAS, RAF, JUN kinase, MYC, and STAT. The end product is leukemogenesis created via a cytokine-independent cell cycle and aberrant apoptotic signals.3
The progression of CML into accelerated phase or blastic phase is associated with the acquisition of genetic or epigenetic abnormalities in particular somatic mutations in genes of chromatin modification of DNA methylation in addition to BCR-ABL rearrangement.4
TET-2 has pleiotropic roles during hematopoiesis, including stem cells self-renewal, lineage commitment, and terminal differentiation of monocytes.5 The chromosome 4q24 region containing theTET-2 gene6 TheTET-2 gene has 11 exons,7 and the resulting messenger RNA (mRNA) may form three different isoforms due to alternative splicing.8 TET 2 is part of the non-driver genetic alterations that may influence myeloproliferative neoplasms development and outcome in general.9
The Ten – Eleven Translocation TET proteins TET-1,TET-2, TET-3 are α - ketoglutarate and Fe2+ dependent enzymes capable of modifying DNA methylation status.10 TET family enzymes and 5- hmC are critical in epigenetic regulation during development.11 Homozygous and heterozygous mutations in TET-2 gene are recurrent events in human hematopoietic malignancies. Most of these mutations decreaseTET-2 enzymatic activity by truncating the protein or affecting its catalytic activity. TET-2 deletion is sufficient to initiate myeloid and lymphoid transformation, including CML.12 The role of TET2 polymorphism is not fully established according to the prognostic and responsiveness to treatment in the context of myeloid malignancies, mainly CML.
Aim of Work
The aim of this study is to evaluate the incidence of the TET2 single nucleotide polymorphism (SNP) (rs2454206, rs34402524, rs61744960) in chronic myeloid leukemia and healthy controls in relation to the disease’ prognostic-criteria.
Materials and Method
The study included 84 subjects; 54 cases were diagnosed as CML, and 30 subjects as a control group matched for age and sex. Cases were selected from Alexandria Main University Hospital Internal Medicine Department (Hematology Unit) Egypt to determine the selected SNPs genotype frequency. Written Informed consent was taken from every patient and approval of the Ethical committee (IRB No. 00008699, FWA No.00015712) was provided. Routine investigations, including CBC, bone marrow aspiration, biochemical investigations, and molecular study, were performed according to CML to identify the disease stage. A 3ml blood collected via EDTA tubes from either peripheral blood or bone marrow aspirate was done. DNA extraction was performed using Invitrogen purelink genomic DNA minikit (Cat No. k1820-01, Lot No 1510617). The SNP assay for TET2 gene polymorphism, performed with (Thermo-Fisher predesigned SNP, USA) PCR prism 7500 device, included tree polymorphisms that are c-25996719-10, rs 34402524, lot p161221-001 H06, PN*40 (intron missense) c-11566753-20, rs 2454206, lot p161221-001 H08 PN *40 (intron missense) c-25746528, rs 61744960, lot p161221-001 H05 PN*40 (intron missense).
Patients were started on Imatinib 400mg per oral daily after confirming the diagnosis and were followed up monthly by CBC, liver and renal function tests as a routine for filling up their prescriptions. At 3 and 6 months, BCR ABL1 IS% was done by real time PCR to monitor response during which no dose adjustments were required, and mild side effects were reported in the form of mild musculoskeletal pain treated by paracetamol.
Statistical analysis
The software of IBM SPSS 20 was used. Data were tested for normality using the Kolmogorov-Smirnov test, Shapiro-Wilk test. Measurement data were displayed in the form of minimum, maximum, mean ± SD. T-test was used for comparing means in parametric data. Qualitative data were displayed in percentages and tested by Pearson’s Chi Square and Fisher Exact Test according to the categories and cells estimation %. If the distribution was nonparametric in distribution, measurement data were displayed in the form of median value and range, and the nonparametric test (Mann-Whitney U) was used for comparing median value. Spearman bivariate correlation analysis was used for analyzing correlation. P<0.05 indicated statistical significance.
Results
The mean age was 45.98±15.7 yrs in CML patients and 39.3±6.587 yrs in the control group (p>0.05). TET2 SNP rs 34402524 was either heterozygous or homozygous in CML (48%, and 46.2% respectively) but was mainly homozygous among the control (80%) group (p=0.012). TET2 SNP rs 2454206 wild type within CML was 65.4% and in control was 63.3% group (p=0.046) (Table 1, Figure 1) TET2 SNP rs 61744960 showed a homozygous pattern among all groups (CML and control) (p=0.528). CML counts were not influenced by TET2 polymorphism, including Hb level, platelets count, WBC, basophil count, and bone marrow blasts (P>0.05) (Table 2).TET2 SNP in CML cases did not alter the prognostic criteria as no statistical significance was noted (p>0.05) except for TET2 SNP rs3442524 homozygous group that was significantly related to huge spleen size with homozygous group and higher BCR-ABL1 levels after six months of starting TKIs (p<0.05) (Table 3).
Table 1.
TET2 single polymorphism patterns for the 3 positions among the three studied groups (CML, and control).
| TET2 SNP | CML | Control | Test of significance | |||
|---|---|---|---|---|---|---|
|
| ||||||
| No. (n=54) | % | No. (n=30) | % | |||
|
| ||||||
| rs 34402524* | Wild | 3 | 5.8 | 0 | 0 | F=0.677 P=0.826 |
| Heterozygous | 25 | 48.0 | 6 | 20 | ||
| Homozygous | 24 | 46.2 | 24 | 80 | ||
|
| ||||||
| rs 2454206* | Wild | 34 | 65.4 | 19 | 63.3 | X2 =0.491 P=0.483 |
| Heterozygous | 18 | 34.6 | 11 | 36.7 | ||
| Homozygous | 0 | 0 | 0 | 0 | ||
|
| ||||||
| rs 61744960* | Heterozygous | 2 | 3.8 | 0 | 0 | - |
| Homozygous | 50 | 96.2 | 30 | 100 | ||
F=fisher exact test, X2 chi square, P<0.05 considered significant,
2 cases failure.
Figure 1.
Distribution of Single Nucleotide Polymorphism Among the Studied Group.
Table 2.
Comparison between prognostic factors in CML cases in chronic phase and TET2 polymorphism.
| SNP | CML | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| No. | Age (yrs) | Hb (g/dl) | Platelets (count/cmm) | Basophils (%) | Marrow Blasts (%) | Spleen size (cm) | WBC (count/cmm) | Sokal score | |
|
| |||||||||
| Median | Mean + SD | Median | Median | Median | Mean ±SD | Median | Mean ±SD | ||
|
| |||||||||
| Rs 34402524 | |||||||||
| Wild | 3 | 52 | 10.7+1 | 390000 | 6 | 1 | 18.75±3.2 | 16000 79000 66500 | 1.01±0.057 |
| Heterozygous | 25 | 50 | 10.39+1.98 | 458000 | 2 | 2 | 20.4±3.6 | 1.33±0.28 | |
| Homozygous | 24 | 40 | 9.61+2.3 | 298000 | 4 | 2 | 24.3±4.1 | 1.45±0.26 | |
|
| |||||||||
| Test of significance | KW=2.64 P=0.267 |
P=0.47* | KW=5.53 P=0.63 |
KW=2.82 P=0.244 |
KW=4.02 P=0.134 |
p=0.019* | KW=5.27, p=0.07 | p=0.87* | |
|
| |||||||||
| Rs 2454206 | |||||||||
| Wild | 34 | 40 | 10.22+2.11 | 323000 | 3 | 2 | 21.47±4.41 | 67000 70000 | 1.31±0.29 |
| Heterozygous | 18 | 50 | 9.71+2.1 | 293000 | 4 | 2 | 24.05±3.68 | 1.44±0.25 | |
|
| |||||||||
| Test of significance | MW=234 P=0.421 |
P=0.481* | MW= 130 P=0.317 |
MW=277 P=0.771 |
MW=179 P=0.627 |
p=0.108* | KW=1.478, P=0.224 | p=0.221* | |
|
| |||||||||
| Rs 61744960 | |||||||||
| Heterozygous | 2 | 50 | 10.4+0.5 | 308000 | 8 | 1 | 23.5+1.5 | 455500 67000 | 1.4 1.366±0.28 |
| Homozygous | 50 | 47 | 10.035+2.11 | 309000 | 4 | 2 | 22.27±4.34 | ||
|
| |||||||||
| Test of significance | P=0.84** | P=0.866** | P=0.964** | P=0.823** | P=0.261** | p=0.54* | KW= 2.265, p= 0.132 | p=0.907* | |
p <0.05 considered significant.
tested by anova. kw kruskal wallis,
mw mann whitney.
Table 3.
Comparison between tet2 polymorphism and BCR ABL1 IS% in chronic phase CML.
| SNP | CML BCR-ABL1 IS% level | ||
|---|---|---|---|
|
| |||
| No. | Initially | 6 months | |
|
| |||
| Median | Median | ||
|
| |||
| Rs 34402524 | |||
| Wild | 3 | 88.3+20.2 | 0.37+0.548 |
| Heterozygous | 25 | 100 | 6.4+15.194 |
| Homozygous | 24 | 99.32+4.85 | 23.2 +28.469 |
|
| |||
| Test of significance | KW=16.33 P=0.000 |
KW=8.315 P=0.016 |
|
|
| |||
| Rs 2454206 | |||
| Wild | 34 | 98.97+6 | 11.82+24.26 |
| Heterozygous | 18 | 100 | 17.55+22.24 |
|
| |||
| Test of significance | P=0.467** | P=0.529** | |
|
| |||
| Rs 61744960 | |||
| Heterozygous | 2 | 100 | 0.76+0.339 |
| Homozygous | 50 | 99.3+4.94 | 14.33+23.846 |
|
| |||
| Test of significance | P=0.98** | P=0.763** | |
p <0.05 considered significant. KW Kruskal wallis,
MW Mann whitney.
Discussion
TET2 regulates epigenetics modification of DNA via regulation of cytosine methylation, and that is crucial for stem cells and progenitor cells’ self-renewal and leukemia prevention. TET2 mutations are common in hematological diseases, including CML. TET2 polymorphism is influenced by racial variation. Little was reported on the influence of TET2 polymorphism among myeloid neoplasms. Studies linked acute leukemia to various TET2 polymorphism influencing the outcome, and this might suggest a role of TET2 polymorphism and its mutations in transforming CML from chronic phase to accelerated phase or blast crisis.
In our study, TET2 SNP rs 34402524 among chronic phase CML was either heterozygous 48% or homozygous 46% but was mainly homozygous among the control group (80%) (p=0.012). As for TET2 SNP rs 2454206, wild type in CML and control group were (62.5%, 63.3% respectively) (p=0.046). TET2 SNP rs 61744960 showed a homozygous pattern among all groups (CML 95.8% and control 100%) (p=0.528).
The studied TET2 SNP might be present as part of a germline phenotype influenced by racial background, as noted within the control group. Variations of TET2 SNP between the control group and CML may suggest that CML patients may have lacked or lost these germline phenotypes during leukomogenesis. The three profiles of TET2 and BCR-ABL emergence are possible: TET2 could proceed BCR-ABL in these cases all Ph positive and Ph negative are TET2 mutant. A biclonal disease, in this case, all Ph positive cells are TET2 wild type. BCR-ABL could occur before TET2 mutation suggesting a late diagnosis of CML.2
Kutny et al., in 201513 in a study involving 403 patients enrolled in Children’s Cancer Group reported that the 10 SNPs with higher prevalence (4%–54%), only the most prevalent SNP, rs2454206 (A>G, I1762V) was associated with survival. OS was significantly higher for patients with minor allele genotypes (TET2 AG/GG) than those with TET2 AA genotype (60±10% vs. 38±11% at 5 years, log-rank P=0.013 TET2, the genotype was influenced by racial variations. TET2AA genotype occurred in 79% of black vs. 39% of white patients (p<0.001). However, both in black and withe people, the survival was higher in TET2 AG/GG respect to TET AA genotype.
Li et al. from Taiwan in 201114 reported that about 78.6% of patients were diagnosed with TET2 SNP (single nucleotide polymorphism). All SNPs were heterozygous, and only 4 SNP were homozygous; all were in SNP rs2454206 (I1762V).14
In our study, as for chronic phase CML, TET2 SNP rs 34402524 homozygous group was significantly related to larger spleen size and BCR-ABL1 IS% levels at initial diagnosis and after six months on follow up after starting TKIs. This suggests that it may play a role in disease progression within Egyptian population (p<0.05). The other TET2 polymorphisms were common too, yet they did not alter the prognostic criteria for CML patients regarding the Sokal score, WBC count, and spleen size (p>0.05).
The present paper is the first to relate TET2 polymorphism to prognostic parameters, and further studies, within the Egyptian population.
Conclusions and Recommendations
TET2 SNP is a common finding among Egyptian chronic myeloid leukemia patients. TET2 SNP rs 34402524 was associated with huge spleen size and higher BCR-ABL1 levels after six months of starting TKIs denoting the possibility of its relation to disease progression.
Ethical standards
All procedures in this research was subjected to the ethical regulation of ethical committee and a Written Informed consent was taken from every patient and approval of the Ethical committee of the Faculty Of Medicine–Alexandria University according to ICH GCP guidelines and applicable local and institutional regulations and guidelines governing EC operation (IRB No. 00008699, FWA No.00015712) full-filling the 1964 Helsinki declaration.
Footnotes
Competing interests: The authors declare no conflict of Interest.
References
- 1.Keramatinia A, Ahadi A, Akbari ME, Mohseny M, Jarahi AM, Mehrvar N, et al. Genomic Profiling of Chronic Myelogenous Leukemia: Basic and Clinical Approach. Journal of Cancer Prevention. 2017;22(2):74–81. doi: 10.15430/JCP.2017.22.2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roche-Lestienne C, Marceau A, Labis E, Nibourel O, Coiteux V, Guilhot J, et al. Mutation analysis of TET2, IDH1, IDH2 and ASXL1 in chronic myeloid leukemia. Leukemia. 2011;25(10):1661–4. doi: 10.1038/leu.2011.139. [DOI] [PubMed] [Google Scholar]
- 3.Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2018 update on diagnosis, therapy and monitoring. American Journal of Hematology. 2018;93(3):442–59. doi: 10.1002/ajh.25011. [DOI] [PubMed] [Google Scholar]
- 4.Kim T, Tyndel MS, Kim HJ, Ahn J-S, Choi SH, Park HJ, et al. Spectrum of somatic mutation dynamics in chronic myeloid leukemia following tyrosine kinase inhibitor therapy. Blood. 2017;129(1):38. doi: 10.1182/blood-2016-04-708560. [DOI] [PubMed] [Google Scholar]
- 5.Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia. 2013:1–36. doi: 10.1038/leu.2013.337. [DOI] [PubMed] [Google Scholar]
- 6.Ko M, Rao A. TET2: epigenetic safeguard for HSC. Blood. 2011;118(17):4501–3. doi: 10.1182/blood-2011-08-373357. [DOI] [PubMed] [Google Scholar]
- 7.Albano F, Anelli L, Zagaria A, Coccaro N, Minervini A, Rossi AR, et al. Decreased TET2 gene expression during chronic myeloid leukemia progression. Leukemia Research. 2011;35(11):e220–e2. doi: 10.1016/j.leukres.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 8.Mohr F, Döhner K, Buske C, Rawat VPS. TET Genes: new players in DNA demethylation and important determinants for stemness. Experimental Hematology. 2011;39(3):272–81. doi: 10.1016/j.exphem.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Shammo JM, Stein BL. Mutations in MPNs: prognostic implications, window to biology, and impact on treatment decisions. Hematology American Society of Hematology Education Program. 2016;2016(1):552–60. doi: 10.1182/asheducation-2016.1.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences. 2011;108(35):14566–71. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia. 2014;28(3):485–96. doi: 10.1038/leu.2013.337. [DOI] [PubMed] [Google Scholar]
- 13.Kutny MA, Alonzo TA, Gamazon ER, Gerbing RB, Geraghty D, Lange B, et al. Ethnic variation of TET2 SNP rs2454206 and association with clinical outcome in childhood AML: a report from the Children/’s Oncology Group. Leukemia. 2015;29(12):2424–6. doi: 10.1038/leu.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M-J, Yang Y-L, Jou S-T, Lu M-Y, Chang H-H, Lin K-H, et al. Prevalence & Prognosis Value of TET2 Gene Polymorphisms in Childhood Acute Myeloid Leukemia in Taiwan. Blood. 2011;118(21):1551. doi: 10.1182/blood.V118.21.1551.1551. [DOI] [Google Scholar]