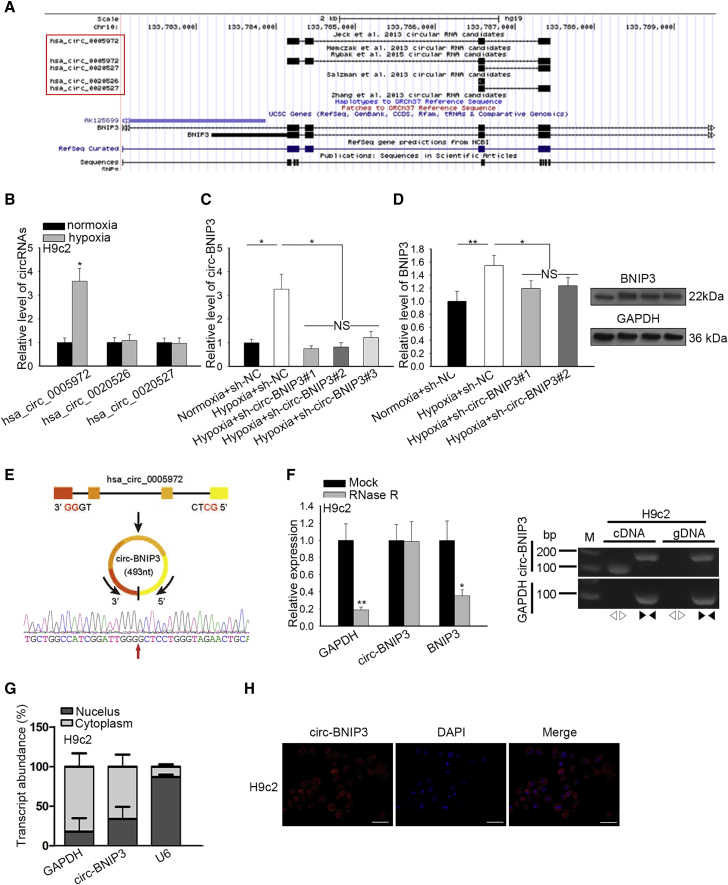
Figure 3.
circ-BNIP3 Was Upregulated by Hypoxia and Positively Regulated BNIP3 in H9c2 Cells
(A) Three circRNAs (hsa_circ_0005972, hsa_circ_0020526, and hsa_circ_0020527) transcribed from BNIP3 were obtained from circBase. The hsa_circ_0005972 was re-termed as circ-BNIP3. (B) Expression of the abovementioned miRNAs was evaluated in hypoxic and normoxic H9c2 cells by quantitative real-time PCR. (C) H9c2 cells underwent hypoxia treatment, with normoxia as control, and were transfected with sh-NC and sh-circ-BNIP3#1/2/3, respectively, under hypoxia. Quantitative real-time PCR analysis for circ-BNIP3 expression in H9c2 cells of each group. (D) Quantitative real-time PCR and western blot determined BNIP3 expression in H9c2 cells of each group. (E) The schematic diagram of formation of circ-BNIP3 and the circRNA sequencing results. (F) Left: relative RNA levels were examined by quantitative real-time PCR and normalized to the value determined in the mock group. Right: circ-BNIP3 amplified by divergent primers were in cRNA rather than in gDNA. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was linear control. (G and H) Circ-BNIP3 was found to be enriched in the cytoplasm of H9c2 cells according to the data of subcellular fractionation assay and FISH (scale bar = 100 μm). Bar graphs were shown as mean ± SD. *p < 0.05, **p < 0.01. NS: no significance.