Abstract
Background
Structured diagnosis (DX) are crucial for secondary use of electronic health record (EHR) data. However, they are often suboptimally recorded. Our previous work showed initial evidence of variable DX recording patterns in oncology charts even after biopsy records are available.
Objective
We verified this finding’s internal and external validity. We hypothesized that this recording pattern would be preserved in a larger cohort of patients for the same disease. We also hypothesized that this effect would vary across subspecialties.
Methods
We extracted DX data from EHRs of patients treated for brain, lung, and pancreatic neoplasms, identified through clinician-led chart reviews. We used statistical methods (i.e., binomial and mixed model regressions) to test our hypotheses.
Results
We found variable recording patterns in brain neoplasm DX (i.e., larger number of distinct DX—OR = 2.2, P < 0.0001, higher descriptive specificity scores—OR = 1.4, P < 0.0001—and much higher entropy after the BX—OR = 3.8 P = 0.004 and OR = 8.0, P < 0.0001), confirming our initial findings. We also found strikingly different patterns for lung and pancreas DX. Although both seemed to have much lower DX sequence entropy after the BX—OR = 0.198, P = 0.015 and OR = 0.099, P = 0.015, respectively compared to OR = 3.8 P = 0.004). We also found statistically significant differences between the brain dataset and both the lung (P < 0.0001) and pancreas (0.009<P < 0.08).
Conclusion
Our results suggest that disease-specific DX entry patterns exist and are established differently by clinical subspecialty. These differences should be accounted for during clinical data reuse and data quality assessments but also during EHR entry system design to maximize accurate, precise and consistent data entry likelihood.
Keywords: data quality, electronic health records, secondary use of clinical data, clinical data management, learning healthcare system
INTRODUCTION
Reliable secondary use of Electronic Health Record (EHR) data is fundamental to learning healthcare systems;1,2 Promising research endeavors such as comparative effectiveness research3,4 and precision medicine5–7 heavily rely on identifying correct patients. Correspondingly, patient Diagnosis (DX) data is often a starting point of cohort selection for secondary use.8,9 Thus, the accurate and precise assignment of structured DX data within EHRs is crucial to ensure reliable research outcomes.10 DX code recording has well-known limitations11 and decades of research have revealed poor DX code data quality through alarmingly high inaccuracy rates.10,12,13 Despite some improvements on error rates over time (i.e., ICD code inaccuracy rates went from 20–70% in the 1970s to 20% in 1980s), their reliability remains questioned.3,14 The complex nature of clinical knowledge, variability of clinical workflows, and clinical data being generated for care and billing rather than research3,15,16 have led to development of workaround methods17–19 including natural language processing (NLP) to access clinical notes, EHR phenotyping, and post hoc data quality assessments.19–24 Though these methods are helpful in unlocking existing poor-quality data, they do not tackle the problem of producing more reliable datasets for future research. Other fields have successfully leveraged data quality improvement techniques ensuring reliable data recording at the source17,21,25,26 but this is generally seen as unfit for the clinical setting due to its complex sociotechnical nature.27,28 These secondary use challenges are compounded by EHR systems that provide multiple descriptions for each individual DX code.29 Though they are intended to facilitate DX code search in practice, the large number of options complicates selecting the most appropriate textual descriptions, increasing the potential erratic DX data recording.30
Although this is a healthcare-wide issue, the problem is compounded and most evident in electronic oncology patient charts.31,32 On one hand, cancer care is team-based and requires multiple specialties and units (e.g., scheduling, encounters, billing, diagnostic imaging, surgical procedures, radiotherapy, etc.); This creates more opportunities for discordant DX recording. EHR systems rarely address this condition, failing to support consistent DX code recording across workflows and making DX logging burdensome to oncologists.33–35 On the other hand, current versions of standard DX codes and descriptions are not designed to support secondary use of clinical data.3 For example, ICD-10 codes C71.* correspond to a malignant neoplasm of the brain DX. These codes allow encoding of the neoplasm site (e.g., C71.1 represents a malignant neoplasm of the frontal lobe) but, as opposed to ICD-O-3 codes,36 they are not designed to provide information on neoplasm type (e.g., IDH mutant glioma, IDH wild type glioma, glioblastoma, etc.) crucial patient classification to treatment selection. However, EHR vendor-provided DX descriptions may include neoplasm type information but vary widely in precision. This complicates structured DX data recording,30 leading to unreliable recording of these DX descriptions preventing their use in patient cohort development for reliable reuse of clinical data.
Our previous work shows that recoding presumably-accurate diagnostic information (i.e., a cancer patient’s biopsy [BX]) does not increase subsequent data accuracy and does not reduce DX data variability in EHRs of patients treated for brain neoplasms in a system with multiple textual DX descriptions per DX.30,37 These findings led us to believe that DX data quality may be affected by the EHR data entry system design and clinical workflows, given that DX data is directly dictated by the BX report in such clinical context. These results also imply that, in the current state of EHR systems design, accurate information does not propagate to populate the rest of the patient’s chart. This is likely to be the case for other segments of the EHR and other clinical specialties. However, we analyzed data for a limited pilot cohort of patients from a single clinical subspecialty. In this study, we tested our initial result’s internal validity of in a larger cohort, hypothesizing that (I) the effect found in the pilot cohort will be preserved in the larger cohort. We also explored their external validity by replicating the study in two other patient cohorts for other oncology subspecialties. Given that DX logging across specialties can vary widely, (this is often noted qualitatively38 but has yet to be shown explicitly and quantitatively in the field) we also hypothesized that (II) DX recording patterns will differ across subspecialties. We tested our hypotheses on EHR data from patients diagnosed with brain, lung and pancreatic neoplasms (i.e., ICD-10 diagnosis codes, C71.*, C34.*, and C25.*). We selected these diseases for their large number of textual diagnosis descriptions (i.e., their potential for precise structured DX recording) for a limited list of specific diagnosis codes, the availability of a clinician-generated patient cohort and each patient chart containing a definitive histopathology report stating the most precise DX description possible. We present descriptive statistics and statistical modeling results that show the existence of seemingly stable DX recording patents that appear to be disease and subspecialty-specific. This analysis contributes to our current understanding of DX logging practices and their differences across workflows and subspecialties by providing concrete, quantitative and explicit evidence of such differences and logging practice variability. Our findings provide further insights to enable reliable reuse of DX data reuse but also lay the groundwork for the development of adaptive clinical data entry support interventions. The resulting understanding aims to unlock new avenues to overcome current challenges introduced by existing EHR systems designs providing multiple DX description per DX code. We also seek to introduce the idea of using such structured textual DX descriptions as a potential source of semi-structured data for cohort selection. Particular care needs to be given to care team and subspecialty differences when making secondary use of clinical data, carrying out clinical data quality assessments and developing patient cohorts using DX data. Our work also suggests support must be provided to clinicians to ensure systematic recording of structured clinical data across subspecialties to enhance clinical data quality and ensure the reliability of secondary analyses of clinical data as a step towards building learning healthcare systems.
METHODS
We extracted diagnoses codes for encounter, problem list, and orders with their corresponding diagnosis descriptions, encounter dates, and ICD-10 codes from the Wake Forest Baptist Medical Center’s Translational Data Warehouse. This database contains all raw data generated during patient care. Our dataset also included surgical pathology reports and relevant covariates such as BX date. We used a clinician-generated gold standard to identify patients treated for each neoplasm. We employed a combination of statistical and NLP tools to quantify DX logging variability in each subspecialty. We built binomial regressions to predict whether DX data appeared before or after the BX based on the number of distinct DX descriptions and DX description “particularity” (i.e., how much semantic information is included in the textual description to differentiate each DX). A “descriptive specificity” or “particularity” score was calculated by extracting clinical concepts from DX descriptions using NLP tools and assigning scores for tumor histology description precision and tumor location.30 Last, we investigated particularity score entropy (i.e., a measure of diversity representing the average amount of information in scores sequences), as a measure of variability in DX descriptions chosen by users before and after the BX. Our study was approved by Wake Forest University School of Medicine’s Institutional Review Board before any data extraction or analysis.
Our patient lists were distilled from the charts of patients treated for brain, lung, and pancreatic neoplasms. Patients were pre-selected during previous clinician-initiated chart reviews. A comprehensive medical record review was performed by two physicians for each patient. The primary post-operative diagnosis was determined based on review of pathology report and progress notes. All treating clinicians were available for consultation when needed. Discrepancies between the two reviewers were resolved by an independent specialist. Our final datasets spanned between January 1st, 2016 to June 1st, 2018. This time frame ensured ICD coding version consistency (i.e., to include DXs after October 2015; ICD-10 implementation date). Each dataset contained 21,136 DX observations for 120 brain neoplasm patients, 119,411 DX observations for 256 lung neoplasm patients and 30,020 DX observations for 34 pancreatic neoplasm patients.
We augmented our dataset with a DX “particularity” score30 based on NLP-extracted39 NCI thesaurus terminology40 concepts from textual DX code descriptions. We define particularity as the ability of a diagnosis description to pinpoint a diagnosis with the most specific concepts available in a relevant controlled clinical vocabulary and provide the most additional descriptive elements (e.g., tumor site) to classify patients. For this study, we used a particularity scoring system to quantify particularity based on both concept specificity and additional descriptive elements. EHRs provide clinicians with these clinically-relevant labels to facilitate DX code selection (i.e., each DX description was linked to an ICD-10 code; C71.*, C34.*, or C25.* codes in our dataset). For example, “Oligodendroglioma of frontal lobe” was attached to C71.1, generically named “Malignant neoplasm of frontal lobe” in the ICD-10 taxonomy. Scores depended on (1) the “descriptiveness” (i.e., relative concept “depth” in the NCI thesaurus structure)40 of the extracted neoplastic process concepts (e.g., glioma is a general neoplastic process description with relative score of “0,” glioblastoma scored “1” as a direct child of the glioma concept in the NCI thesaurus) and (2) whether we found a concept describing the neoplasm’s anatomical site. For example, particularity score in brain DX ranged from 0 for “Malignant neoplasm of brain, unspecified location” to 3 for “Oligodendroglioma of frontal lobe”. We also calculated DX description sequence and DX particularity score entropy as a measure of DX description diversity and DX particularity score variation using the ‘entropy’ R package to foster reproducibility.41 This R package automatically estimates empirical entropy form observation sequences (i.e., DX or Particularity score entropy) using a maximum likelihood estimation algorithm. In the rest of this paper we refer to “DX Entropy” as the entropy of DX sequences and “Score Entropy” as the entropy of particularity score sequences before or after the BX.
We built binomial regressions using R’s generalized linear model42 and mixed models (lme4) packages.43 We selected binomial regressions because our dichotomous main outcome variable ‘After BX’ that indicated whether a DX was recorded before or after the BX. Our original hypotheses30 aimed to explore differences before and after the BX. We evaluated differences in number of distinct DX with a binomial regression model predicting the “After DX” variable based on the number of distinct DX descriptions. The same kind of model was used to estimate differences in DX sequences and particularity score entropy44 values. We built a binomial mixed regression model to evaluate the relationship between DX particularity scores before and after BX. We chose a mixed model to account for each DX score as an independent test and account for intra-subject correlation by attributing random intercepts to each patient. We evaluated the quantitative effect difference across subspecialties by aggregating all three datasets and building the same models controlling for the type of disease diagnosed. We tested for model improvement through covariate inclusion. We included variables such as number of days before or after the BX each DX was recorded, the provider recording the DX and the department (i.e., care units involved in patient treatment such as oncology, surgery, and neurology) where the DX was recorded. We also tested for variable interactions in all models with more than one variable. Summary statistics such as mean, median, and extreme values were employed to screen the data for outliers, missing values and erroneous input. Dates were also reviewed for potential errors such as values being outside the study’s time window. Statistical significance was set at p = 0.05 for all models and adjustments for multiple comparison were made using R’s p.adjust function45 using Holm’s correction method.46 We refer to the odds ratios as OR and adjusted p values as adj-p.
Multiple software tools were used to carry out this analysis. Data extraction and preprocessing was done using a DataGrip software client (version 2017.2.2, JetBrains s.r.o., Prague, Czech Republic). Visual exploration and exploratory descriptive statistics were done using Tableau (version 10.2.4, Tableau Software, Inc., Seattle, WA). All statistical analyses and data manipulation such as data scrubbing and reshaping were done in R version 3.4.130 and RStudio (version 1.1.383, RStudio, Inc., Boston, MA).
RESULTS
Our final datasets contained 21,136 encounter, problem list and order DX observations out of which 2,690 were primary encounter DX for 120 brain neoplasm patients, 119,411 DX observations for 256 lung neoplasm patients, and 30,020 DX observations for 34 pancreatic neoplasm patients (Table 1). Primary encounter brain DX contained 68 distinct DX descriptions corresponding to 8 ICD-10 codes, logged by 144 distinct providers from 47 distinct hospital departments. The average number of days from the BX was 170 ± 189 days. The average particularity score was 0.96 ± 0.71. These statistics were mostly driven by the post-BX data, as expected due to additional treatment data generation. Pre-BX data contained 200 DX observations for 30 patients with 31 distinct DX descriptions corresponding to 4 ICD-10 codes. The number of providers and departments also dipped to 31 and 12 correspondingly. This is expected due to the reduced clinical evidence to make accurate a precise DX before the BX is available as well as the lower number of visits before the treatment plan is defined. Days from BX and Particularity score kept comparable standard deviations, while showing lower averages before the BX as expected (i.e., −137 ± 139 and 200 ± 164 for days and 0.78 ± 0.70 and 0.97 ± 0.71 for pre and post BX particularity). The full datasets containing encounter, problem list, and order DX observations 71,209 and 29 distinct DX descriptions for brain, lung, and pancreas correspondingly. These corresponded to 8, 22, and 10 ICD-10 codes, logged by 268, 592, and 29 distinct providers from 111, 193, and 99 distinct hospital departments. The average number of days from the BX was 189 ± 186, 215 ± 254, and 189 ± 299 days, correspondingly. The average particularity score was 1.01 ± 0.68, 3.76 ± 3.34 and 8.24 ± 9.7, correspondingly, hinting at fundamental differences across DX logging patterns across subspecialties.
Table 1.
Descriptive statistics for primary brain DX and all DX data for brain, lung, and pancreas. These descriptive statistics show differences in DX logging volumes, DX description variability, and DX particularity
| Primary brain DX |
Encounter, problem list, and order DX |
|||||
|---|---|---|---|---|---|---|
| Measure | Overall | Before BX | After BX | Brain | Lung | Pancreas |
| Distinct patients | 120 | 30 | 117 | 120 | 256 | 34 |
| Number of DX records | 2,690 | 200 | 2,490 | 21,136 | 119,411 | 30,020 |
| Distinct ICD-10 codes | 8 | 4 | 8 | 8 | 22 | 10 |
| Distinct DX descriptions | 68 | 28 | 64 | 71 | 209 | 29 |
| Distinct providers | 144 | 31 | 137 | 268 | 592 | 305 |
| Distinct hospital department | 47 | 12 | 46 | 111 | 193 | 99 |
| Days from BX (Mean ±Std.Dev.) | 170 ±189 | 137 ±139 | 200 ±164 | 189 ±186 | 215 ±254 | 189 ±299 |
| DX particularity Score (Mean ±Std.Dev.) | 0.96 ±0.71 | 0.78 ±0.70 | 0.97 ±0.71 | 1.01 ±0.68 | 3.76 ±3.34 | 8.24 ±9.7 |
Our full brain neoplasm DX dataset yielded similar effect sizes the initial results30 for both the primary encounter DX and multi-workflow DX data (Table 2 and Supplementary appendixTable A1). We provide full regression tables in Supplementary Appendix A. For the primary encounter DX dataset, we found that adding an additional distinct DX would make a DX sequence 77.7% more likely to appear after the BX (adj-P = 0.02). Max days from BX was a significant covariate but had a very small effect (OR = 1.004, adj-P = 0.021). For all brain DX, we found that adding a new distinct DX would make a DX sequence 2.2 times more likely to happen after the BX (adj-P < 0.0001). Only problem list DX were statistically different from other DX types (adj-P < 0.0001) (Supplementary Table A1). Particularity score increments of a unit would make a DX 40% more likely to appear after the DX (adj-P < 0.0001); both order and problem list DX were different in this regard (adj-p = 0.02 and adj-P = 0.003). Entropy regressions showed that higher DX sequence entropy made a sequence more likely to happen after the BX (OR = 3.79, adj-P = 0.004) and that higher particularity score also made the sequence more likely to be post-BX (OR = 7.89, adj-P < 0.0001). Neither regression showed any statistically significant covariate relationship after P-adjustment nor any difference across DX types.
Table 2.
Brain DX variability regressions. Post-BX data shows more distinct DX, higher particularity scores but also higher entropy in DX and particularity score sequences
| Model | Term | Odds ratio (exp(ß)) | Confidence interval (95%) | P-value | Adjusted P-value | |
|---|---|---|---|---|---|---|
| Distinct primary DX | Distinct DX number | 1.777 | 0.180 | 1.023 | 0.007 | 0.020 |
| Max days from BX | 1.004 | 0.001 | 0.007 | 0.013 | 0.021 | |
| Distinct DX | Distinct DX number | 2.219 | 0.573 | 1.046 | <0.0001 | <0.001 |
| Particularity | Particularity score | 1.402 | 0.228 | 0.450 | <0.0001 | <0.0001 |
| DX entropy | DX entropy | 3.788 | 0.586 | 2.110 | 0.001 | 0.004 |
| Particularity entropy | Particularity entropy | 7.981 | 1.080 | 3.130 | <0.0001 | <0.0001 |
We found qualitatively different results for both our lung and pancreatic neoplasm DX datasets (Table 3, Supplementary Table A2 and A3). Lung neoplasm revealed a smaller effect in the number of distinct DX (OR = 1.50, adj-P < 0.0001) and the same difference in problem list DX (adj-P = 0.0001). The effect of the particularity score was also much smaller (OR = 1.085, adj-P < 0.0001) with the same differences for order and problem list DX (adj-P < 0.0001). However, higher DX sequence and particularity score entropy made a sequence more likely to happen before the BX (OR = 0.51, adj-P = 0.001, and OR = 0.47, adj-P = 0.017). This means that, contrary to brain neoplasm DX, lung neoplasm DX were less erratic after the BX. Neither of these regressions revealed differences across DX types. None of the lung regressions showed significant covariate relationships or interactions (Supplementary Table A2). Pancreatic neoplasm DX regressions (Table 3 and Supplementary Table A3) revealed no statistical relationship between the number of distinct DX (OR = 1.36, adj-P = 1) nor differences across DX types (adj-P = 1). The effect of the particularity score was opposite (OR = 0.92, adj-P < 0.0001) with DX type differences for order and non-primary encounter DX (adj-P = 0.025 and adj-P = 0.033) rather than problem list DX (adj-P = 0.931). This means that for pancreatic neoplasm DX, a more particular DX description is 9% less likely to appear after the BX. However, higher DX sequence entropy made a sequence more likely to happen before the BX, revealing more consistent logging after the BX (OR = 0.32, adj-P = 0.046). In contrast with brain neoplasm DX and just like lung neoplasm, DX sequences were less erratic after the BX. Particularity score entropy showed no statistically significant relationship (adj-P = 1). Both regressions showed a significant relationship between Post-BX and the number of days from BX but with a very small effect (OR = 1.003, adj-P < 0.0001 for both regressions). Neither regression revealed differences across DX types. No interaction term was found significant in the pancreatic neoplasm regressions.
Table 3.
Lung and pancreatic neoplasm regressions. The effects found for these subspecialties are qualitatively different from those found for brain neoplasm. Both subspecialties seem to present lower entropy values after the BX, in contrast with brain neoplasm DX. The effect of particularity seems different for all three subspecialties
| Sub-specialty | Model & term | Odds ratio (exp(ß)) | Confidence interval (95%) | P-value | Adjusted P-value | |
|---|---|---|---|---|---|---|
| Lung | Distinct DX number | 1.500 | 0.293 | 0.524 | <0.0001 | <0.0001 |
| Particularity score | 1.085 | 0.069 | 0.094 | <0.0001 | <0.0001 | |
| DX entropy | 0.508 | −1.044 | −0.315 | 0.0002 | 0.001 | |
| Particularity entropy | 0.473 | −1.267 | −0.237 | 0.004 | 0.017 | |
| Pancreas | Distinct DX number | 1.363 | −0.145 | 0.810 | 0.197 | 1 |
| Particularity score | 0.915 | −0.109 | −0.067 | <0.0001 | <0.0001 | |
| DX entropy | 0.324 | −2.077 | −0.205 | 0.017 | 0.046 | |
| Particularity entropy | 0.976 | −1.130 | 1.073 | 0.965 | 1.000 | |
We also found quantitative differences across subspecialty datasets (Table 4). We found statistically significant differences between the number of distinct DX in brain neoplasm DX data and lung neoplasm (OR = 0.54, adj-P < 0.0001) as well as pancreatic neoplasm (OR = 0.58, adj-P = 0.009). We found similar differences for particularity scores with significantly lower odds ratios that hinted at much higher particularity in lung and pancreatic neoplasm data (OR = 0.198, adj-P = 0.015 and OR = 0.099, adj-P = 0.015). Entropy values were also significantly different in all cases except for pancreas particularity score entropy (adj-P = 0.080). The entropy models revealed statistically significant differences hinting at lower entropy in the lung and pancreas datasets as compared to the brain dataset (OR = 0.53, adj-P < 0.0001, and OR = 0.56, adj-P = 0.014 for DX sequence entropy, for lung and pancreas correspondingly; OR = 0.54, adj-P < 0.0001 for lung particularity score entropy). For both entropy regressions, we found a statistically significant relationship with max days from BX with a low effect size (OR = 1.005, adj-P < 0.0001 in both cases). No other covariates were significantly correlated. No interactions were found significant.
Table 4.
Effect comparison regressions. Both lung and pancreatic neoplasm data seem to have quantitatively different effects from brain neoplasm DX in terms of distinct DX number, particularity scores, and entropy
| Model | Term | Odds ratio (exp(ß)) | Confidence interval (95%) | P-value | Adjusted P-value | |
|---|---|---|---|---|---|---|
| Distinct DX | Distinct DX number | 1.457 | 0.292 | 0.464 | <0.0001 | <0.0001 |
| Brain | 1 | – | – | – | – | |
| Lung | 0.538 | −0.911 | −0.333 | <0.0001 | <0.0001 | |
| Pancreas | 0.580 | −0.919 | −0.171 | 0.004 | 0.009 | |
| Particularity | Particularity score | 1.041 | 0.030 | 0.050 | <0.0001 | <0.0001 |
| Brain | 1 | – | – | – | – | |
| Lung | 0.198 | −2.912 | −0.374 | 0.012 | 0.015 | |
| Pancreas | 0.099 | −4.043 | −0.625 | 0.007 | 0.015 | |
| DX Entropy | DX entropy | 0.904 | −0.415 | 0.212 | 0.525 | 1 |
| Max days from BX | 1.005 | 0.004 | 0.005 | <0.0001 | <0.0001 | |
| Brain | 1 | – | – | – | – | |
| Lung | 0.529 | −0.944 | −0.334 | <0.0001 | <0.0001 | |
| Pancreas | 0.561 | −0.980 | −0.178 | 0.005 | 0.014 | |
| Particularity Entropy | Particularity entropy | 1.581 | 0.031 | 0.888 | 0.036 | 0.080 |
| Max days from BX | 1.005 | 0.004 | 0.005 | <0.0001 | <0.0001 | |
| Brain | 1 | – | – | – | – | |
| Lung | 0.542 | −0.923 | −0.308 | <0.0001 | <0.0001 | |
| Pancreas | 0.628 | −0.877 | −0.054 | 0.027 | 0.080 | |
In summary, we confirmed the internal validity of our initial results for brain neoplasm cases (i.e., larger number of distinct DX, higher particularity scores, and higher entropy after the BX); odds ratio values also close. Replicating the analysis in lung and pancreatic neoplasm patient charts, we found DX logging pattern differences across subspecialties. Lung and pancreas DX both seemed to have lower DX sequence entropy and therefore more consistent DX recording after the BX contrarily to brain neoplasm DX. Particularity odds ratios were different for all three datasets; differences between datasets were statistically significant in most cases.
DISCUSSION
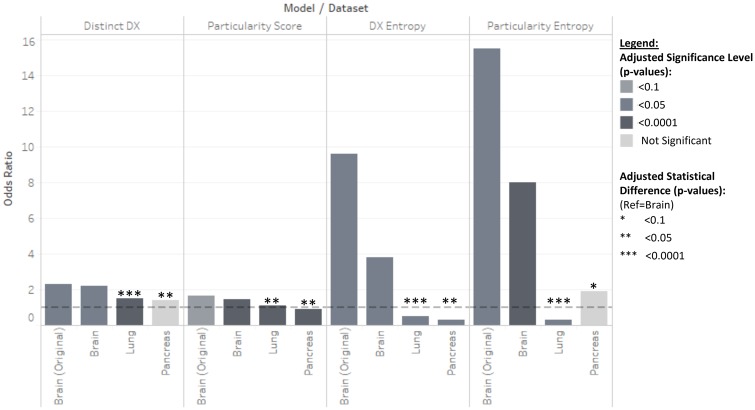
We used statistical regressions to evaluate structured DX data differences before and after BX reports are recorded in EHR records for oncology patients (Figure 1). We found that DX entry patterns were similar for two different brain cancer cohorts but qualitatively and quantitatively different from cohorts of patients treated for lung and pancreatic cancer. These results provide quantitative evidence that suggests that clinicians establish a stable EHR interaction mechanism for entering structured DX data, that this workflow remains consistent over time and that it may vary by subspecialty or disease-type. This implies that interaction design for data entry should be carefully considered by EHR vendors and clinical interface designers to support the accurate and precise data entry. The subspecialty differences have important implications as efforts to improve and optimize DX data entry may thus be different based on the disease area. This hints at the need for disease-process-specific data entry support, which justifies the use of advanced data-driven methods such as machine learning to achieve personalized support for the entry of accurate clinical data. Data quality assessment research and method development efforts should also take these data production differences into account when evaluating a repurposed dataset’s fitness for purpose.47–51
Figure 1.
Odds ratio and statistical difference findings summary.
Our study extends the existing literature by exploring aspects beyond the predominant DX code recording accuracy analyses.1,32,52 We showed the limited impact of a DX source of truth (i.e., the BX report) in the EHR on subsequent DX recording, confirming our previous findings for a pilot cohort of patients treated for brain neoplasms.30 Though previous studies have explored multiple EHR segments to assess data quality,11,13,31,32,38,53 they often focus on providing accuracy measure results to warn secondary users of clinical data against direct analysis of repurposed clinical data. Our study takes this a step further by exploring patterns in a segment of the EHR where no variability should exist due to the availability of a BX that provides a source of accurate information. This was the main reason for choosing oncological charts. We were also unable to find similar analyses on repurposed EHR data quality focusing specifically on oncological DX data. Thus, our study focuses on breaking new ground and opening new avenues of research from the vantage point of oncological patient charts into all segments of EHR databases. It is interesting that we found evidence that DX recording processes may be stable for different disciplines within a common disease state. For example, we collected DX data for specialists in medical oncology, neuro-oncology, radiation oncology, and neurosurgical oncology for patients with primary brain neoplasms. Despite the differences in specialty and training, relatively little variability was observed within this patient cohort likely reflecting the common clinical workflows that are established by clinicians caring for this patient population. In contrast, we observed significant differences between the DX data for this multidisciplinary care team and a similar multidisciplinary lung and pancreatic team. This is congruent with previous accuracy-driven EHR data analyses.38 Our findings stress the ties between clinical practice to the resulting data. Secondary analyses of clinical data1,3,54 and data quality assessments48,50,51,55,56 need to account for such cross-practice differences. Though our analysis was conducted exclusively on oncology EHRs, we used these data as a setup to better understand the impact of clinical data entry.
We also explored the impact of multiple descriptions linked to single diagnostic codes on structured data entry. To our knowledge, only our initial analysis30 has explored the effect of such a setup. Finding that clinicians logged more distinct DX descriptions after the BX in two out of three subspecialties confirms our initial hypothesis that multiple DX description per code may increase DX entry variability even after the DX is known through the BX report. This observation also raises questions about how the currently available DX code lists within the EHR drive clinician practice. Anecdotal discussions with clinicians in these specialty areas suggest that DX data tables available in the EHR may drive some of the clinical practices observed in this study. Subsequent investigation is currently being conducted to better understand this finding. This, in turn, confirms that data entry and the resulting intrinsic data quality49,57 are likely to be impacted by EHR interface design and demanding clinical workflows.33,58,59 This corroborates that clinical data is a tangential product of clinical care and is affected by clinical processes such as billing,34,60 rather than the ideal raw material for clinical research.3 Much current research aiming to support secondary analysis of clinical data aims to develop methods for data quality assessment and data curation in isolation from the clinical practice.4,48–50,55,56,61–64 Yet, our results suggest that the quality of extracted clinical data is directly linked to the process that produced them; They also suggest that data quality can be improved by altering the process. For example, integrating specialty-dependent diagnosis lists (e.g., ICD-O) into the EHR system could be beneficial to improve data entry, quality, and concordance. The data entry process could also be revised. For example, ICD-O diagnoses could be presented first for oncology patients or could be prompted when a pathology report indicates a cancer diagnosis in the EHR. It is crucial that future development in this area should take into account such cross-process differences to ensure the reliable secondary use of clinical data.65
Our analysis presents three main limitations. First, we focused our study on a limited number of clinical conditions (i.e., brain, lung, and pancreatic neoplasms). However, we were able to evaluate differences across subspecialties quantitatively and qualitatively finding different DX logging profiles for each subspecialty. Our data and analyses provided adequate evidence to evaluate our hypotheses and test the internal and external validity of our initial results. We will further extend this analysis to other subspecialties to better understand DX logging differences across clinical workflows; We also expand to specialties outside oncology. A related limitation was not accounting for comorbidities. However, the informatics-driven nature of our analysis, along with the relatively large number of patients for each dataset and large effect sizes suggest that the phenomenon is driven by clinical practice and workflows rather than the patient’s clinical profile. Comorbidity effects will also be explored in future work. Second, our particularity scoring has yet to be validated with a gold standard. However, our DX particularity scoring was simple, transparent, and systematic enough to be considered a repeatable feature extraction process.66,67 The score was based on summing a point for anatomical location concepts found and neoplastic process concept’s precision based on its depth in the NCI Thesaurus classification,40 a well-known and standardized classification of clinical concepts. The validation for this method is planned in our future work. Finally, we only evaluated DX description variability (number of distinct DX), average particularity before and after BX and the entropy of these two values rather than DX accuracy. Although it would have been ideal to evaluate the accuracy of each DX entry as a measure of intrinsic data quality, we defined our variables to measure variability in a context where no variability should exist because the DX is known in the EHR through the BX report. In this context, DX variation is a reliable proxy for inaccuracy that our regressions were able to uncover.
Future work will be divided into three segments: further analyses to confirm DX logging patterns differences across subspecialties, the exploration of root causes through qualitative research and the development of workflow-adaptive interventions to increase DX data quality and support systematic DX logging across specialties. First, we will explore differences across other specialties and validate our particularity scoring system. We will also analyze temporal patterns to better understand data entry dynamics. Then, we will carry out qualitative research such as interviews and focus groups to explore the underlying causes DX logging differences across subspecialties. We will also carry out additional quantitative analyses to explore potential factors influencing data entry processes and contributing to DX data entry variability. Finally, we will employ informatics methods to develop clinician-adaptive interventions to support systematic, accurate and concordant DX recording leveraging fragmented data across EHRs. We will also explore the idea of centralized DX entry to assess whether such intervention increases DX concordance across the EHR.
CONCLUSION
Our analysis provides quantitative evidence showing that disease-specific DX entry patterns exist and are established differently by clinical subspecialty. Secondary users of clinical data should consider these differences when designing analyses and performing data quality assessments. Standardized interventions able to accommodate these differences and support systematic, accurate, precise, and concordant DX entry across subspecialties must be developed and implemented to increase the reliability of structured DX data and enable reliable secondary analyses of clinical data. Overcoming such challenges may support the improvement of overall clinical data quality,48,49,57 reliability of secondary analyses of clinical data1 and the building of the learning system.31,68,69
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
FUNDING STATEMENT
This work was supported by the Cancer Center Support Grant from the National Cancer Institute to the Comprehensive Cancer Center of Wake Forest Baptist Medical Center (P30 CA012197), by the National Institute of General Medical Sciences’ Institutional Research and Academic Career Development Award (IRACDA) program (K12-GM102773) and by Wake Forest Baptist Health’s Center for Biomedical Informatics’ Pilot Award.
COMPETING INTERESTS STATEMENT
The authors have no competing interests to declare.
CONTRIBUTOR STATEMENT
Dr. Diaz-Garelli was responsible for the conception, design, data extraction, analysis and writing of the paper as well as revision of the majority of the paper and final approval. Dr. Strowd contributed to the acquisition of clinician-reviewed brain biopsy data and provided critical feedback for data analysis and result interpretation. Dr. Ahmed contributed to the acquisition of clinician-reviewed lung biopsy data and provided critical feedback for data analysis and result interpretation. Dr. Wells provided feedback on statistical methodology and analysis design. Dr. Pasche contributed to the acquisition of clinician-reviewed pancreatic biopsy data and provided critical feedback for data analysis and result interpretation. Dr. Laurini contributed to the interpretation of our results from the clinical pathology point of view. Ms. Merrill contributed to the acquisition of clinician-reviewed brain biopsy data. Dr. Topaloglu contributed to the conception of this research and paper. He also provided major feedback on the methods and paper design. All authors helped revise the entire paper and gave approval of the final version. The authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge use of the services and facilities, funded by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (UL1TR001420). We would also like to acknowledge Thomas Lycan Jr., D.O. and Andrew Dothard, M.D. as well as Riddhishkumar Shah M.D., John Migliano M.D. and Sandrine Crane M.D. who for their contribution in curating the lung and pancreatic neoplasm datasets respectively.
Conflict of interest statement. None declared.
REFERENCES
- 1. Safran C. Reuse of clinical data. Yearb Med Inform 2014; 9: 52–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Safran C. Using routinely collected data for clinical research. Stat Med 1991; 104: 559–64. [DOI] [PubMed] [Google Scholar]
- 3. Hersh WR, Weiner MG, Embi PJ, et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care 2013; 51: S30–S37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown JS, Kahn M, Toh D.. Data quality assessment for comparative effectiveness research in distributed data networks. Med Care 2013; 51: S22–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beckmann JS, Lew D.. Reconciling evidence-based medicine and precision medicine in the era of big data: challenges and opportunities. Genome Med 2016; 8: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chambers DA, Feero WG, Khoury MJ.. Convergence of implementation science, precision medicine, and the learning health care system. A new model for biomedical research. JAMA 2016; 315: 1941–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. National Research Council (US) Committee on a Framework for Developing a New Taxonomy of Disease. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC:National Academies Press; 2011. [PubMed] [Google Scholar]
- 8. Köpcke F, Prokosch H-U.. Employing computers for the recruitment into clinical trials: a comprehensive systematic review. J Med Internet Res 2014; 16. doi: 10.2196/jmir.3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moskowitz A, Chen K.. Defining the patient cohort In: Secondary Analysis of Electronic Health Records. Springer International Publishing; 2016: 93–100. doi: 10.1007/978-3-319-43742-2_10 [Google Scholar]
- 10. Doremus HD, Michenzi EM.. Data quality: an illustration of its potential impact upon a diagnosis-related group’s case mix index and reimbursement. Med Care 1983; 2110: 1001–11. [PubMed] [Google Scholar]
- 11. Hsia DC, Krushat WM, Fagan AB, et al. Accuracy of diagnostic coding for medicare patients under the prospective-payment system. 10.1056/NEJM198802113180604. 2010. doi: 10.1056/NEJ M1 9 8 802113180604; Last accessed April 21, 2019. [DOI] [PubMed]
- 12. Lloyd SS, Rissing JP.. Physician and coding errors in patient records. JAMA 1985; 25410: 1330–6. [PubMed] [Google Scholar]
- 13. Johnson AN, Appel GL.. DRGs and hospital case records: implications for Medicare case mix accuracy. Inquiry 1984; 212: 128–34. [PubMed] [Google Scholar]
- 14. Botsis T, Hartvigsen G, Chen F, et al. Secondary use of EHR: data quality issues and informatics opportunities. AMIA Summit Translat Sci Proc 2010; 2010: 1.. [PMC free article] [PubMed] [Google Scholar]
- 15. Blois MS. Information and Medicine: The Nature of Medical Descriptions. Oakland, CA: University of California Press; 1984. [Google Scholar]
- 16. Sacchi L, Dagliati A, Bellazzi R.. Analyzing complex patients’ temporal histories: new frontiers in temporal data mining In: Fernández-Llatas C, García-Gómez JM, eds. Data Mining in Clinical Medicine. New York: Springer; 2015: 89–105. [DOI] [PubMed] [Google Scholar]
- 17. Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of electronic health record oriented phenotyping algorithms. AMIA Annu Symp Proc 2011; 2011: 274–83. [PMC free article] [PubMed] [Google Scholar]
- 18. Wei W-Q, Teixeira PL, Mo H, et al. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc 2016; 23 (e1): e20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sarmiento RF, Dernoncourt F.. Improving patient cohort identification using natural language processing In: Secondary Analysis of Electronic Health Records. Springer International Publishing; 2016: 405–17. doi: 10.1007/978-3-319-43742-2_28 [PubMed] [Google Scholar]
- 20. Burger G, Abu-Hanna A, de Keizer N, et al. Natural language processing in pathology: a scoping review. J Clin Pathol 2016; 6911: 949–55. [DOI] [PubMed] [Google Scholar]
- 21. Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004; 115: 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dagliati A, Sacchi L, Zambelli A, et al. Temporal electronic phenotyping by mining careflows of breast cancer patients. J Biomed Inform 2017; 66: 136–47. [DOI] [PubMed] [Google Scholar]
- 23. Halpern Y. Semi-supervised learning for electronic phenotyping in support of precision medicine. 2016. http://search.proquest.com/docview/1848662728/abstract/CBCE93B15C7A498BPQ/1 Accessed April 6, 2017.
- 24. Hripcsak G, Albers DJ.. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 2013; 201: 117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Batini C, Scannapieca M.. Methodologies for data quality measurement and improvement In: Data Quality: Concepts, Methodologies and Techniques. Springer Berlin Heidelberg; 2006: 161–200. http://link.springer.com/chapter/10.1007/3-540-33173-5_7 (accessed 31 Jul 2014). [Google Scholar]
- 26. Khalil OEM, Harcar TD.. Relationship marketing and data quality management. SAM Adv Manag J Corpus Christi 1999; 64: 26–33. [Google Scholar]
- 27. Ash JS, Sittig DF, Campbell EM, et al. Some unintended consequences of clinical decision support systems. AMIA Annu Symp Proc 2007; 2007: 26–30. [PMC free article] [PubMed] [Google Scholar]
- 28. Campbell EM, Sittig DF, Guappone KP, et al. Overdependence on technology: an unintended adverse consequence of computerized provider order entry. AMIA Annu Symp Proc 2007; 2007: 94–8. [PMC free article] [PubMed] [Google Scholar]
- 29. Baskaran L, Greco PJ, Kaelber DC.. Case report medical eponyms. Appl Clin Inform 2012; 3: 349–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diaz-Garelli J-F, Wells BJ, Yelton C, et al. Biopsy records do not reduce diagnosis variability in cancer patient EHRs: are we more uncertain after knowing? AMIA Jt Summits Transl Sci Proc 2018; 2017: 72–80. [PMC free article] [PubMed] [Google Scholar]
- 31. Farzandipour M, Sheikhtaheri A, Sadoughi F.. Effective factors on accuracy of principal diagnosis coding based on International Classification of Diseases, the 10th revision (ICD-10). Int J Inform Manag 2010; 301: 78–84. [Google Scholar]
- 32. O’Malley KJ, Cook KF, Price MD, et al. Measuring diagnoses: ICD code accuracy. Health Serv Res 2005; 40: 1620–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walji MF, Kalenderian E, Tran D, et al. Detection and characterization of usability problems in structured data entry interfaces in dentistry. Int J Med Inform 2013; 822: 128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Howard J, Clark EC, Friedman A, et al. Electronic health record impact on work burden in small, unaffiliated, community-based primary care practices. J Gen Intern Med 2013; 281: 107–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Asan O, Nattinger AB, Gurses AP, et al. Oncologists’ views regarding the role of electronic health records in care coordination. JCO Clin Cancer Inform 2018; 2: 1–12. [DOI] [PubMed] [Google Scholar]
- 36. Jack A, Organization WH, Percy CL, et al. International Classification of Diseases for Oncology: ICD-O. Geneva, Switzerland: World Health Organization; 2000. [Google Scholar]
- 37. Diaz-Garelli J-F, Wells BJ, Merrill R, et al. Lost in Translation: Diagnosis Records Show More Inaccuracies After Biopsy in Oncology Care EHRs San Francisco, CA; 2019. [PMC free article] [PubMed]
- 38. Fleming M, MacFarlane D, Torres WE, et al. Magnitude of impact, overall and on subspecialties, of transitioning in radiology from ICD-9 to ICD-10 codes. J Am Coll Radiol 2015; 1211: 1155–61. [DOI] [PubMed] [Google Scholar]
- 39. Tseytlin E, Mitchell K, Legowski E, et al. NOBLE – Flexible concept recognition for large-scale biomedical natural language processing. BMC Bioinformatics 2016; 17. doi: 10.1186/s12859-015-0871-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sioutos N, Coronado S. D, Haber MW, et al. NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information. J Biomed Inform 2007; 401: 30–43. [DOI] [PubMed] [Google Scholar]
- 41. Hausser J, Strimmer K. Entropy: estimation of entropy, mutual information and related quantities. 2014. https://cran.r-project.org/web/packages/entropy/index.html; Last accessed April 21, 2019.
- 42. R Core Team. R: A Language and Environment for Statistical Computing Vienna, Austria: R Foundation for Statistical Computing; 2013.
- 43. Bates D, Maechler M, Bolker B, et al. lme4: linear mixed-effects models using “Eigen” and S4. 2017. https://cran.r-project.org/web/packages/lme4/index.html; Last accessed April 21, 2019.
- 44. Shenkin PS, Erman B, Mastrandrea LD.. Information-theoretical entropy as a measure of sequence variability. Proteins 1991; 114: 297–313. [DOI] [PubMed] [Google Scholar]
- 45. Wright SP. Adjusted P-values for simultaneous inference. Biometrics 1992; 484: 1005. [Google Scholar]
- 46. Aickin M, Gensler H.. Adjusting for multiple testing when reporting research results: the Bonferroni vs Holm methods. Am J Public Health 1996; 865: 726–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Holve E, Kahn M, Nahm M, et al. A comprehensive framework for data quality assessment in CER. AMIA Jt Summits Transl Sci Proc 2013; 2013: 86–8. [PMC free article] [PubMed] [Google Scholar]
- 48. Kahn MG, Callahan TJ, Barnard J, et al. A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. EGEMS (Wash DC) 2016; 4. doi: 10.13063/2327-9214.1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weiskopf NG, Weng C.. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc 2013; 201: 144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kahn M, Brown J, Chun A, et al. Transparent reporting of data quality in distributed data networks. EGEMS (Wash DC) 2015; 3. doi: 10.13063/2327-9214.1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weiskopf NG, Bakken S, Hripcsak G, et al. A data quality assessment guideline for electronic health record data reuse. EGEMS ((Wash DC) 2017; 5. doi: 10.13063/egems.1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Escudié J-B, Rance B, Malamut G, et al. A novel data-driven workflow combining literature and electronic health records to estimate comorbidities burden for a specific disease: a case study on autoimmune comorbidities in patients with celiac disease. BMC Med Inform Decis Mak 2017; 17: 140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pippenger M, Holloway RG, Vickrey BG.. Neurologists’ use of ICD-9CM codes for dementia. Neurology 2001; 569: 1206–9. [DOI] [PubMed] [Google Scholar]
- 54. Weiner MG, Embi PJ.. Toward reuse of clinical data for research and quality improvement: the end of the beginning? Ann Intern Med 2009; 1515: 359–60. [DOI] [PubMed] [Google Scholar]
- 55. Kahn MG, Raebel MA, Glanz JM, et al. A pragmatic framework for single-site and multisite data quality assessment in electronic health record-based clinical research. Med Care 2012; 50. doi: 10.1097/MLR.0b013e318257dd67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Callahan TJ, Barnard JG, Helmkamp LJ, et al. Reporting data quality assessment results: identifying individual and organizational barriers and solutions. EGEMS (Wash DC) 2017; 5. doi: 10.5334/egems.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang RY, Strong DM.. Beyond accuracy: what data quality means to data consumers. J Manag Inform Syst 1996; 124: 5–33. [Google Scholar]
- 58. Zhang J, Walji MF.. TURF: Toward a unified framework of EHR usability. J Biomed Inform 2011; 446: 1056–67. [DOI] [PubMed] [Google Scholar]
- 59. Abran A, Khelifi A, Suryn W, et al. Usability meanings and interpretations in ISO standards. Softw Qual J 2003; 114: 325–38. [Google Scholar]
- 60. Richesson RL, Horvath MM, Rusincovitch SA.. Clinical research informatics and electronic health record data. Yearb Med Inform 2014; 9: 215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hripcsak G, Duke J, Shah N, et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. MEDINFO 2015; 15. [PMC free article] [PubMed] [Google Scholar]
- 62. Johnson SG, Speedie S, Simon G, et al. A Data Quality Ontology for the Secondary Use of EHR Data. San Francisco, CA: AMIA; 2015. [PMC free article] [PubMed] [Google Scholar]
- 63. Johnson SG, Speedie S, Simon G, et al. Application of an ontology for characterizing data quality for a secondary use of EHR data. Appl Clin Inform 2016; 7: 69–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DQe-v: A database-agnostic framework for exploring variability in electronic health record data across time and site location. https://egems.academyhealth.org/articles/10.13063/2327-9214.1277/ Accessed June 18, 2018. [DOI] [PMC free article] [PubMed]
- 65. Medicine I of, Medicine R on E-B. The Learning Healthcare System: Workshop Summary. Washington, DC: National Academies Press; 2007. [PubMed] [Google Scholar]
- 66. Scott S, Matwin S.. Feature engineering for text classification In: Proceedings of ICML-99, 16th International Conference on Machine Learning. Morgan Kaufmann Publishers; 1999: 379–88. [Google Scholar]
- 67. Blum AL, Langley P.. Selection of relevant features and examples in machine learning. Artif Intell 1997; 97 (1–2): 245–71. [Google Scholar]
- 68. Lorence D. Regional variation in Medical Classification Agreement: benchmarking the coding gap. J Med Syst 2003; 27: 435–43. [DOI] [PubMed] [Google Scholar]
- 69. Burgun A, Botti G, Beux PL.. Issues in the design of medical ontologies used for knowledge sharing. J Med Syst 2001; 25: 95–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.