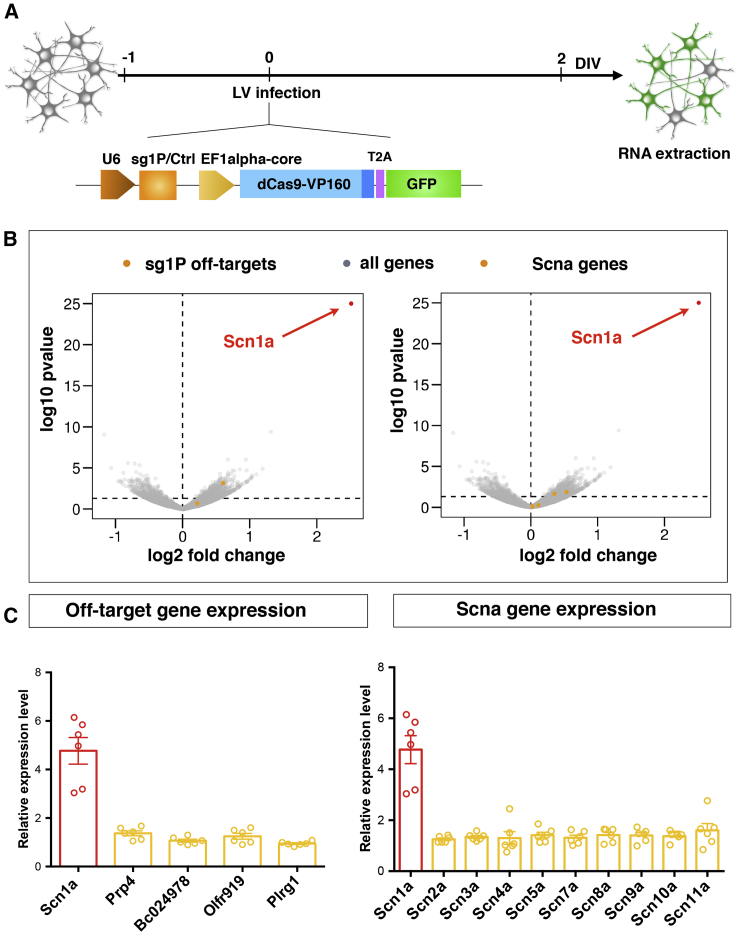
Figure 3.
Global Gene Expression analysis of Transduced Neurons Confirms the High Specificity Profile of the Scn1a-dCas9A System
(A) Schematic view of the experimental setting to perform gene expression profiling of Ctrl-dCas9A- and Scn1a-dCas9A-treated primary neurons. E17.5 embryo-derived neurons were transduced with single LVs at DIV 1 expressing either the Ctrl-dCas9A or Scn1a-dCas9A elements and processed for RNA extraction 48 h later (DIV 3). (B) Volcano plots showing the log10 p value as a function of log2 fold changes in gene expression in Scn1a-dCas9A-treated neurons with respect to Ctrl-dCas9A. Scn1a is shown as a red dot. Yellow dots represent off-target genes in the left panel and other Scna genes in the right panel. All other genes are shown as gray dots. (C) qRT-PCRs for profiling the expression of predicted off-targets (genes Prp4, BC024978, Olfr919, and Plrg1; left panel) or other Scna genes (Scn2a, Scn3a, Scn4a, Scn5a, Scn7a, Scn8a, Scn9a, and Scn11a; right panel). Plotted values are normalized on 18S rRNA and expressed as relative to sgCtrl-treated samples (value = 1, data not shown). n = 6; sg1p versus sgCtrl p < 0.0004, Student’s t test. Data are shown as mean ± SEM, with dots representing individual samples.