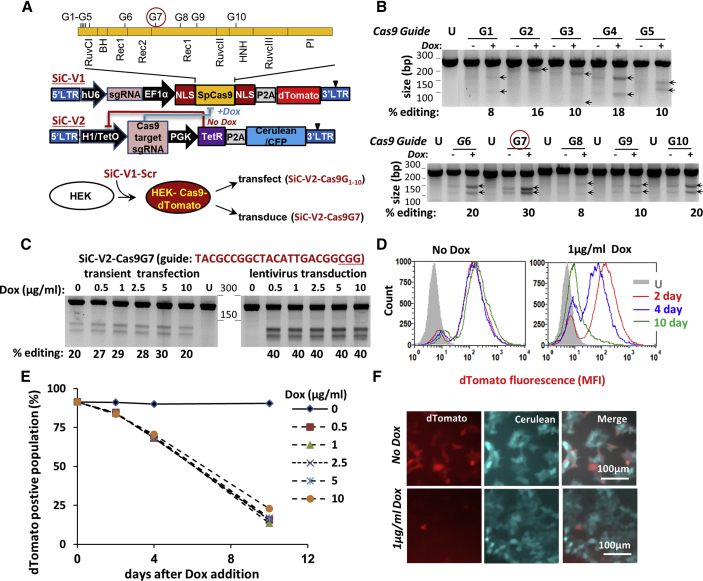
Figure 1.
Self-Inactivating CRISPR-Cas9 System, SiC
(A) Lentiviral vector with two constructs: SiC-V1 and SiC-V2. SgRNA editing Cas9 are numbered G1–G10, along with their site of action (top). Experiment schematic is shown at the bottom. (B) sgRNA G1–G10 were cloned individually into SiC-V2, and transiently transfected into HEK-Cas9dTomato cells, which stably express Cas9 and dTomato. Following 72 h with and without 2.5 μg/mL Dox, the Surveyor assay was performed. Percent of gene editing was quantified using densitometry. “U” indicates untransfected (wild-type) HEK cells. Arrows indicate indels. sgRNA G7 in the Rec1 lobe was most efficient at editing Cas9. Data are representative of 2–4 repeats for each sgRNA. (C) HEK-Cas9dTomato cells were either transiently transfected (left half of gel) or stably transduced (right half) to express SiC-V2-Cas9G7 (SiC-V2 with guide G7). The latter stably transduced cells are called “HEK-Cas9dTomato-Cas9G7. Dox concentration was varied over 72 h prior to Surveyor assay to monitor Cas9 editing. Cas9 editing is strictly Dox-dependent upon lentiviral transduction. Data are representative of 3–4 repeats. (D–F) Time course studies of Cas9 activity performed using “HEK-Cas9dTomato-Cas9G7” cells. dTomato reporter fluorescence measured using cytometry decreased gradually over 10 days in the presence of 1 μg/mL Dox (D). Genome editing rate was independent of Dox concentration in the range tested (0.5–10 μg/mL, E) Microscopy image shows >90% loss of dTomato signal after 10 days of Dox (F). Data are representative of >10 repeats. See also Figures S1 and S2.