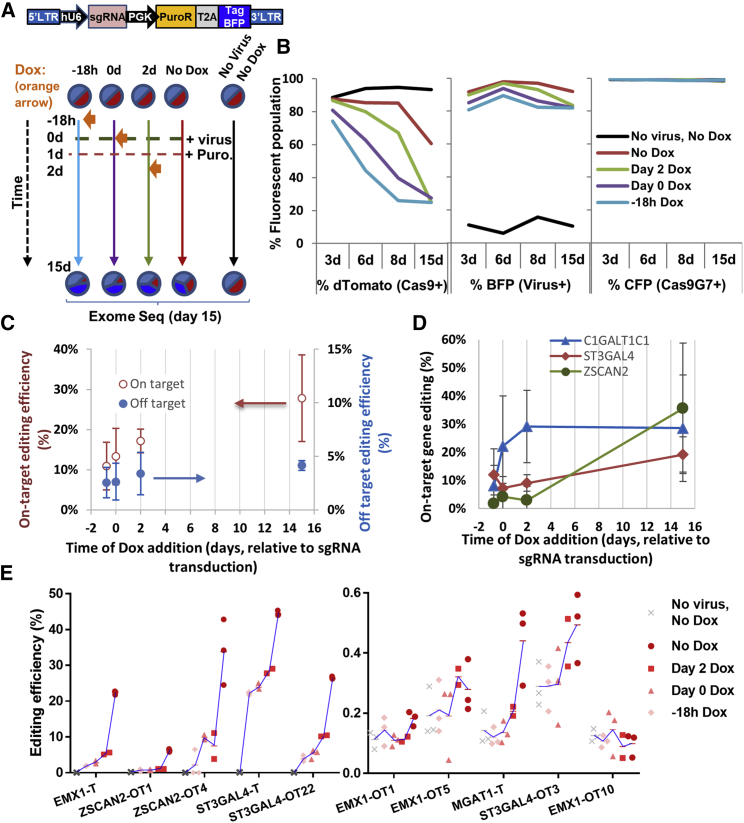
Figure 2.
Time-Dependent On- and Off-Target Editing Assessed Using Exome and Deep Sequencing
(A) HEK-Cas9dTomato-Cas9G7 cells were transduced with a pool of five lentivirus carrying promiscuous sgRNA targeting human EMX1, C1GalT1C1, MGAT1, ST3Gal4, or ZSCAN2. Cells were red due to Cas9dTomato, cyan due to Cas9G7 sgRNA, which was co-expressed with a Cerulean reporter, and blue due to BFP co-expression by the virus carrying the promiscuous sgRNA. 1 μg/mL Dox was added either 18 h prior to viral transduction, at day 0 (0 day) or at day 2 (2 day) in order to self-inactivate Cas9 (A). Dox was absent in one case, and control cells remained untransduced (without Dox). 1 μg/mL puromycin was added to select for transduced cells starting at day 1 (1 day). Genomic DNA was purified at day 15 for exome sequencing (results in C and D). Selected on- and off-target sites were also amplified from genomic DNA, and indels were quantified using deep sequencing (results in E). (B) Cas9 expression (dTomato, red), presence of sgRNA-library (BFP, blue), and Cas9G7 sgRNA (Cerulean, Cyan) expression were monitored at various times using surrogate fluorescence reporters. Note that some decrease in dTomato fluorescence is observed in No Dox sample at day 15 as the cells were cultured to over-confluence, but there is no Dox editing in these cells based on deep-sequencing. (C and D) In exome sequencing runs, data are presented as mean ± SD for indel formation on 3 on-target genes (C1GalT1C1, ST3Gal4, ZSCAN2) and 32 putative off-target genes. Among the 32, 30 are exonic off-target edits and 2 are intronic. Here, both on-target and off-target editing increased if Dox addition was delayed. Low levels of off-target editing (0.27%) and no on-target editing (0%) was detected in the no-virus control sample, and thus the study design contains only low levels of basal noise. In general, off-target editing was ∼10-fold lower compared to on-target (C). (D) On-target editing percent for three genes based on triplicate runs at each time point. Here, whereas some genes were edited at early times, indels accumulated in others over a period of days (D). (E) In deep-sequencing runs, also, on- and off-target editing increased upon delaying Dox addition. Here, “-T” denotes on-target and “-OT” denotes off-target editing. Samples at each time were analyzed in triplicate as noted using red symbols. Samples with lower duration of Dox treatment are indicated using darker red symbols and these appear to the right in the individual plots. Untransduced cells and cells with earlier Dox addition time points appear to the left. A blue line links the mean “%indel values” of each of the samples, starting with “No virus, No Dox,” to earlier Dox to later, No Dox treatment samples. Percent indel was higher in some runs (left panel) compared to others (right panel), with no editing seen in EMX1-OT10. See also Figure S3.