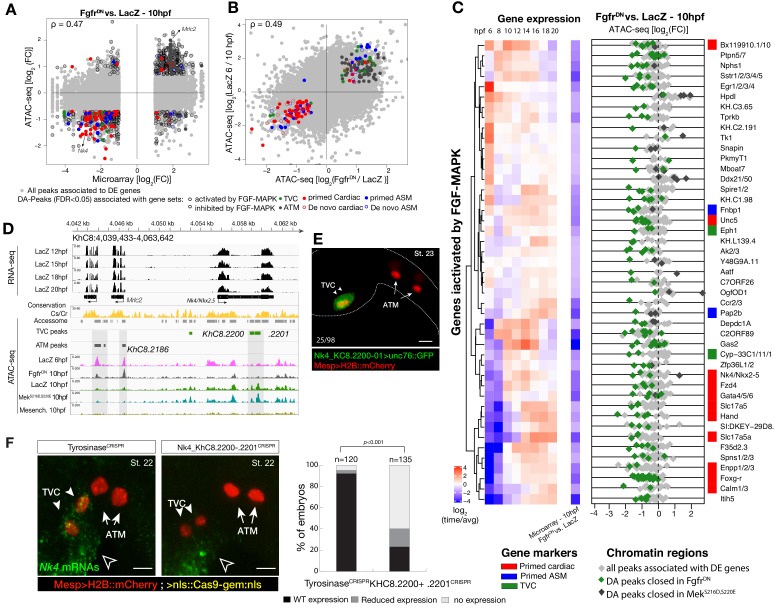
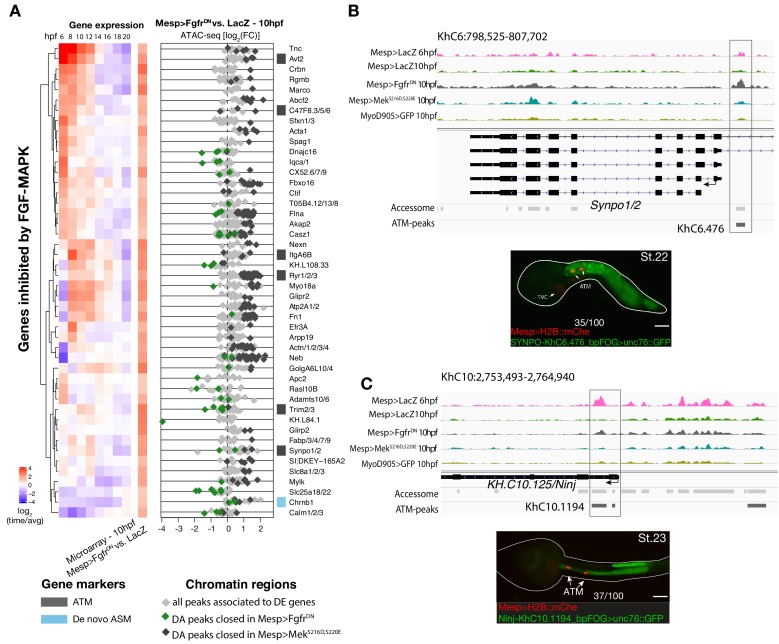
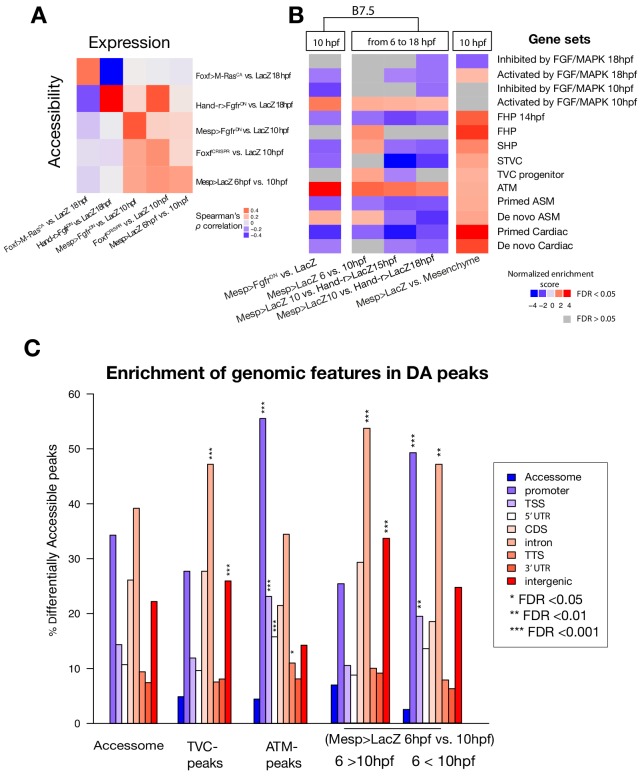
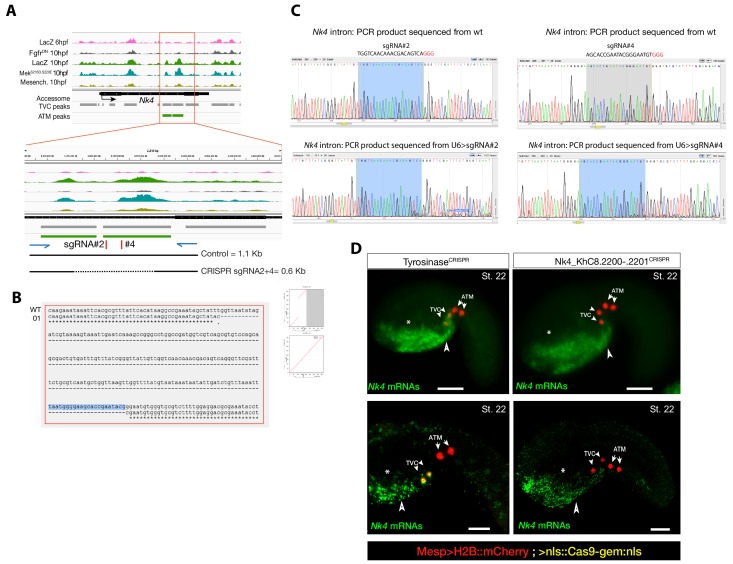
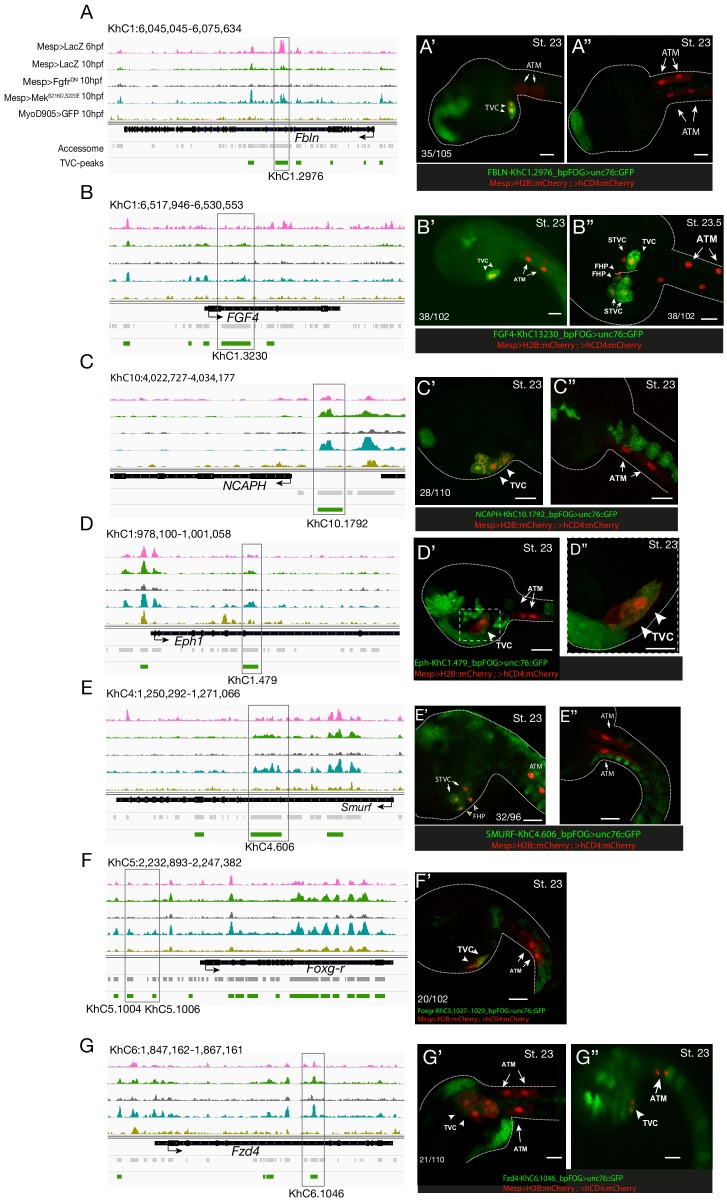
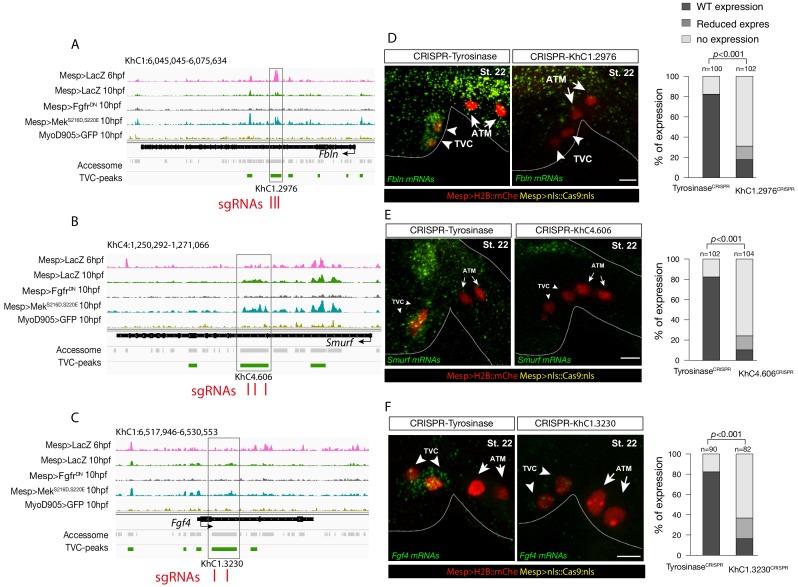
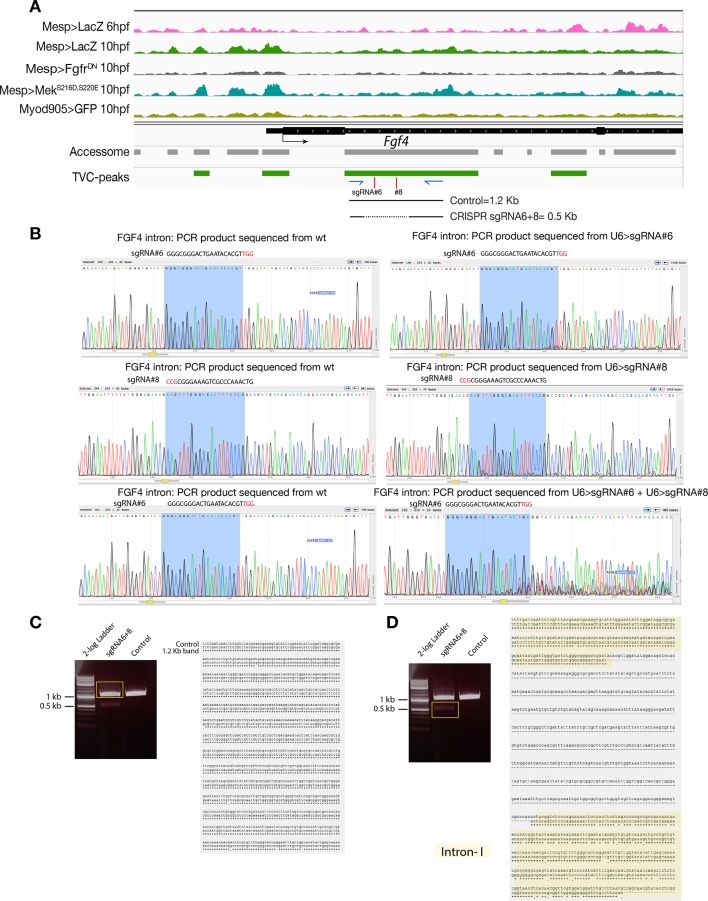
Figure 2. Cardiopharyngeal accessibility profiles are established in multipotent progenitors.
(A–B) Correlations between differential gene expression (DE) and differential chromatin accessibility (DA) in response to FGF-MAPK perturbation in the multipotent progenitors (A) and between chromatin accessibility in response to FGF-MAPK perturbation and in multipotent progenitors (10 hpf) versus founder cells (6 hpf). (B). Colored dots are DA peaks associated with cell type-specific DE genes. ρ is the Spearman correlation of expression and accessibility for DA regions associated with DE genes (A) or of region response to MAPK perturbation with accessibility in founder cells versus multipotent progenitors (B). (C) Relationship between expression and accessibility of DE genes associated with DA regions for genes in the bottom 0.75% quantile of fold change between expression in FgfrDN and control (log2(FC) < −1.32). Microarray log2(fold change (FC)) values are shown on the left. The fold change for all time points is versus the average. (D) A 24 kb region on chromosome eight displaying expression (RNA-seq) and chromatin accessibility (ATAC-seq; normalized by total sequencing depth). Gray shaded boxes show validated ATM-specific promoters and a newly identified TVC-specific enhancer in Nk4/Nkx2-5 intron. (E) Enhancer-driven in vivo reporter expression (green) of tested ATAC-seq regions (KhC8.2200 and .2201). TVCs marked with Mesp>H2B::mCherry (red). Numbers indicate observed/total of half-embryos scored. (F) Endogenous expression of Nk4/Nkx2-5 visualized by in situ (green) in TyrosinaseCRISPR and upon CRISPR/Cas9-induced deletions of TVC-specific region. Nuclei of B7.5 lineage cells are labelled by Mesp>nls::LacZ and revealed with an anti beta-galactosidase antibody (red). Nk4/Nkx2-5 expression was not affected in the epidermis (open arrowhead). Experiment performed in biological replicates. Scale bar, 20 μm. Fisher exact test, total numbers of individual halves scored per condition are shown in 'n='. Gene expression data for 6 hpf and ‘FGF-MAPK perturbation 10 hpf’ (Christiaen et al., 2008) and 8 to 20 hpf (Razy-Krajka et al., 2014) were previously published.