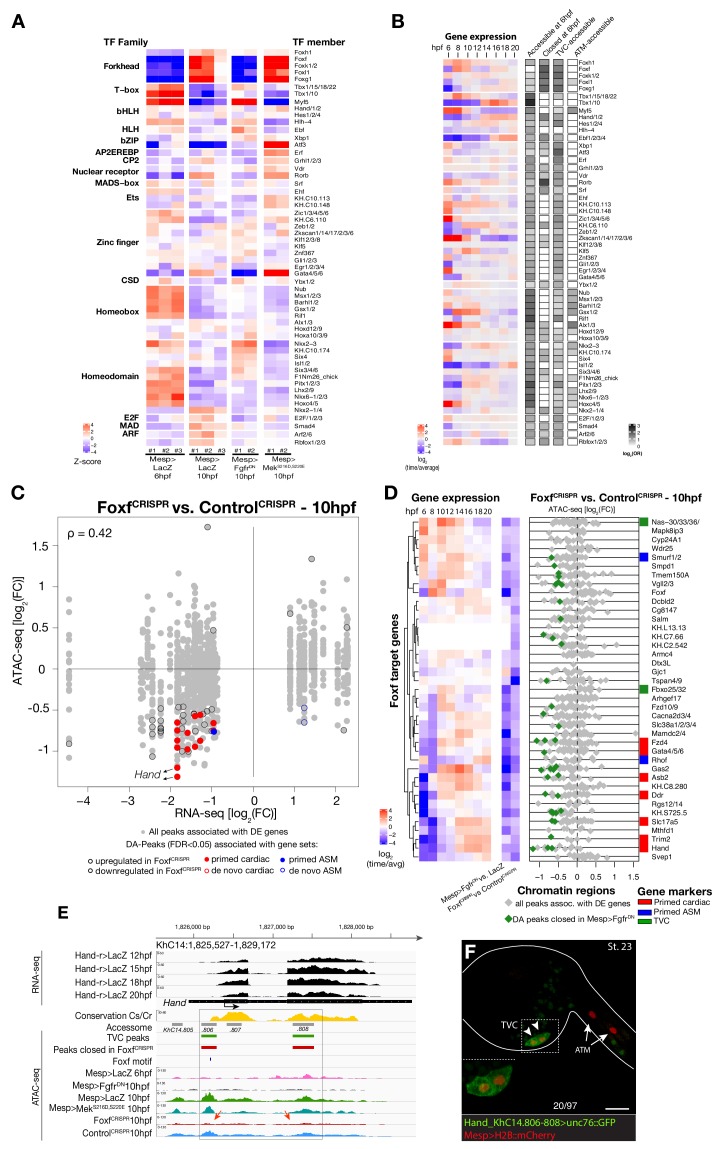
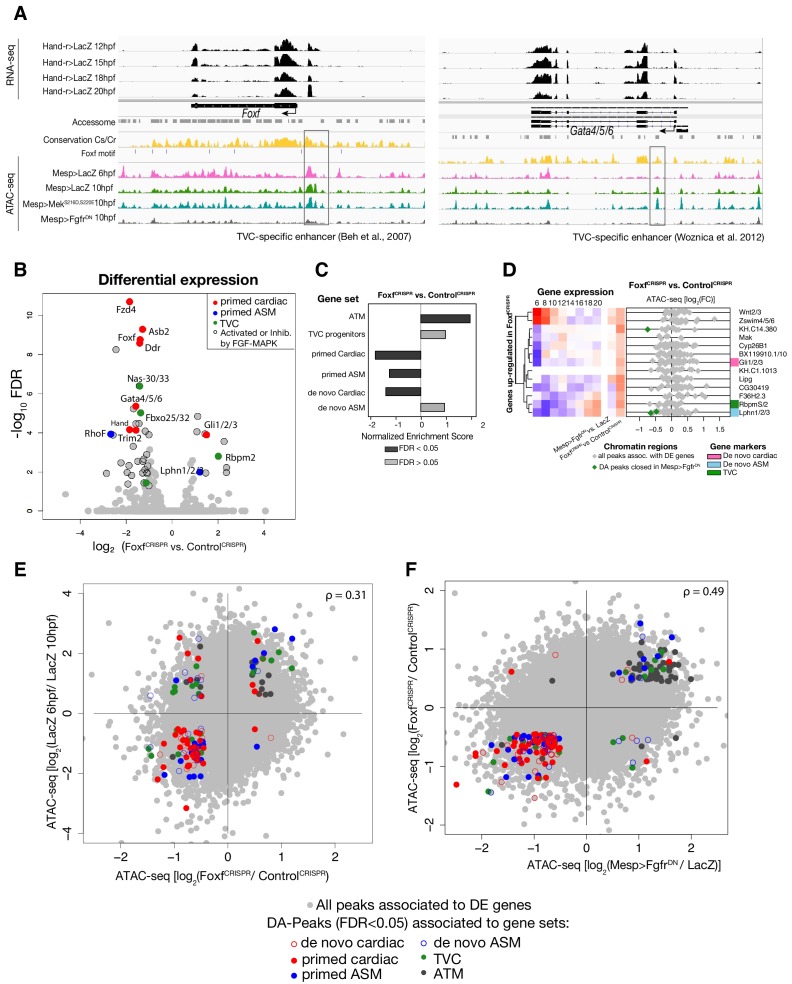
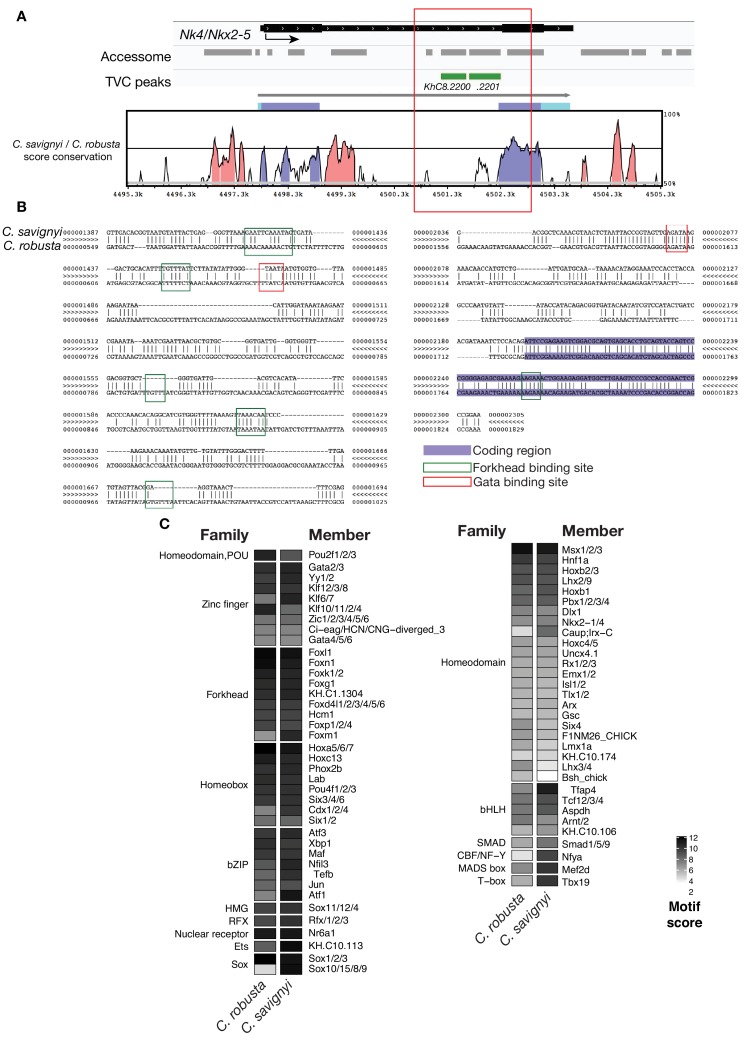
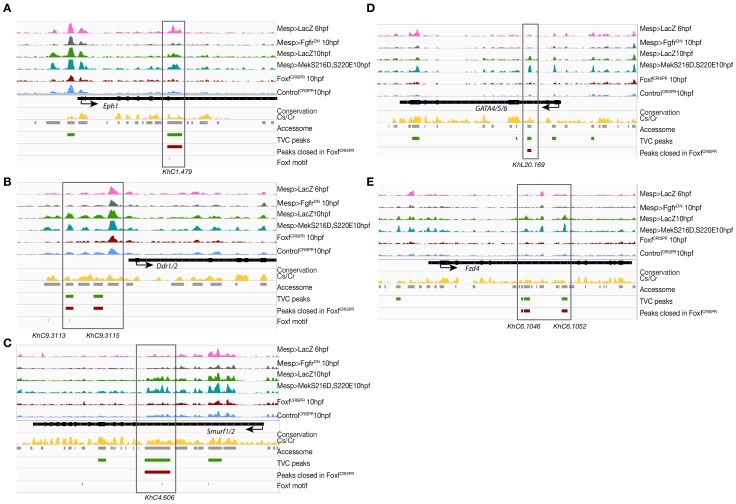
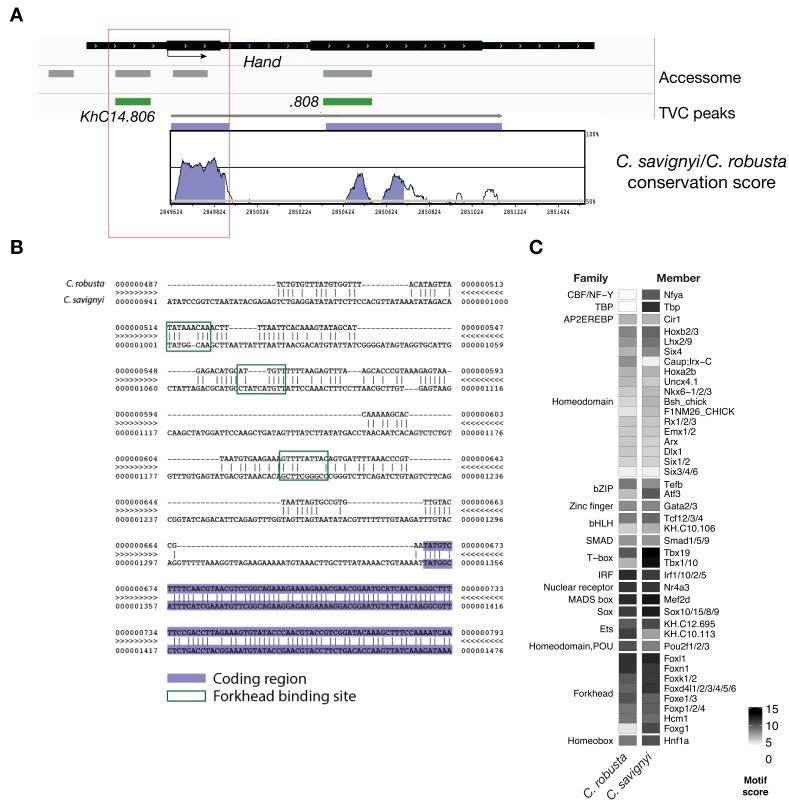
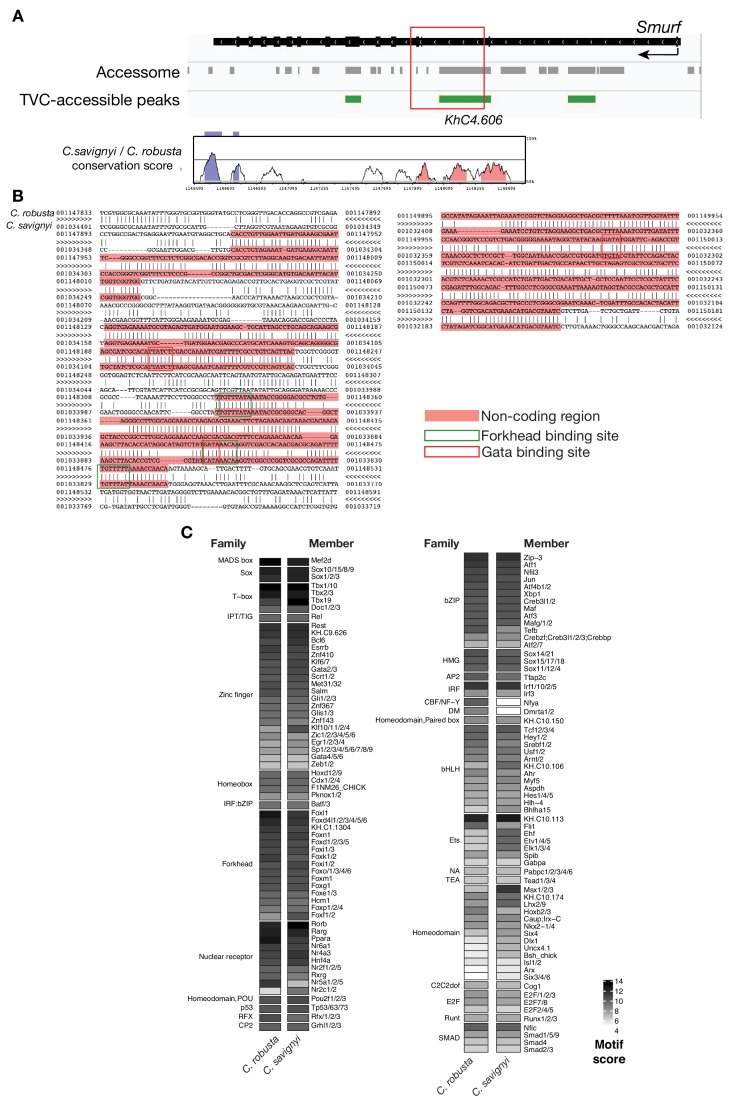
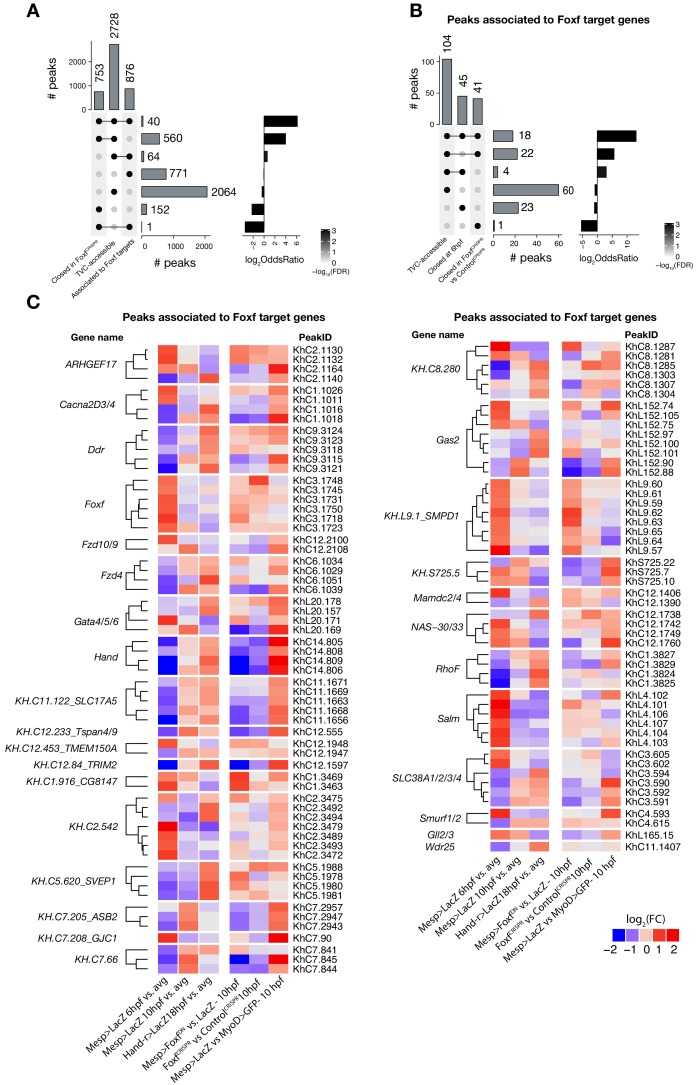
Figure 3. Foxf is required for cardiopharyngeal-specific chromatin accessibility.
(A) Motif accessibility between libraries from chromVAR (Schep et al., 2017). Motifs were obtained and associated with Ciona transcription factors (TFs) as described in the Materials and methods. Deviations were computed for FGF signaling-dependent regions at 10 hpf and B7.5 replicates at 6 and 10 hpf. We calculated the differential accessibility of all motifs between conditions and time points. Only the most significant motif is shown for each TF. (B) Expression of transcription factors over time compared to enrichment of corresponding TF motifs in condition specific peak sets. log2(odds ratio) values (log2(OR), see Materials and methods) are shown for motifs that are significantly enriched in a peak set (one-tailed hypergeometric test, FDR < 0.05). Only TFs expressed in the B7.5 lineage are shown. (C) Differential expression of Foxf target genes (DE) vs. differential chromatin accessibility (DA) in FoxfCRISPR. ρ is the Spearman correlation of expression and accessibility for DA regions associated with DE genes. (D) Association between expression of Foxf target genes and accessibility of proximal regions which were both TVC-specific and closed in FoxfCRISPR as in Figure 2C. (E) A 3.6 kb region on chromosome 14 displaying expression profiles of RNA-seq and chromatin accessibility profiles of ATAC-seq normalized tag count. Foxf core binding site (GTAAACA) is displayed as blue line. The boxed region indicates a newly identified TVC-specific enhancer in Hand locus. Red arrow indicates a TVC-specific enhancer showing closed chromatin in FoxfCRISPR ATAC-seq. (F) Enhancer-driven in vivo reporter expression (green) of tested ATAC-seq peaks. TVCs marked with Mesp>H2B::mCherry (red). Numbers indicate observed/total of half-embryos scored. Experiment performed in biological replicates. Scale bar, 30 μm. Gene expression data for 6 hpf and ‘FGF-MAPK perturbation 10 hpf’ (Christiaen et al., 2008), and from 8 to 20 hpf (Razy-Krajka et al., 2014) were previously published.