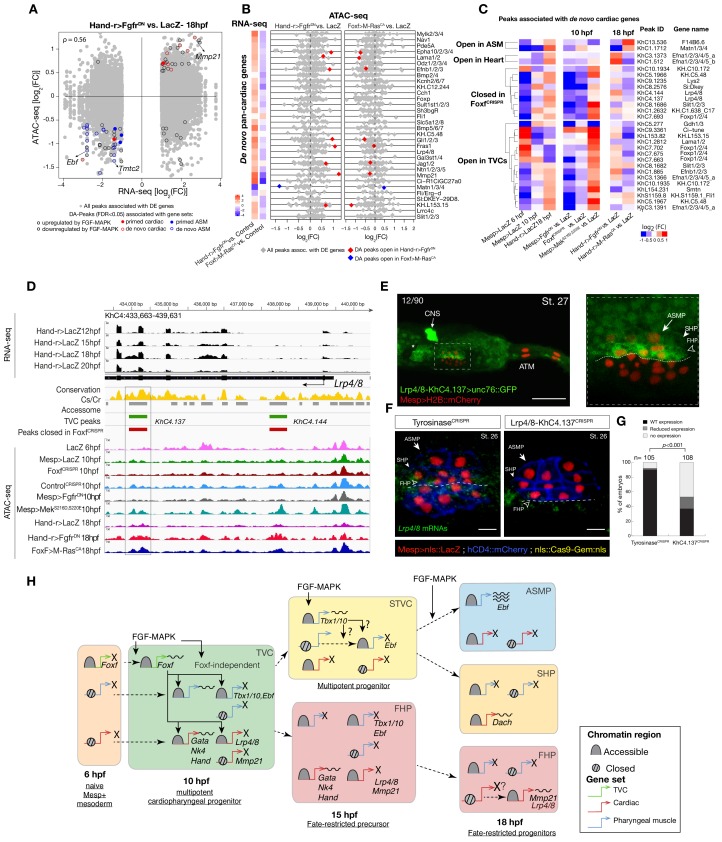
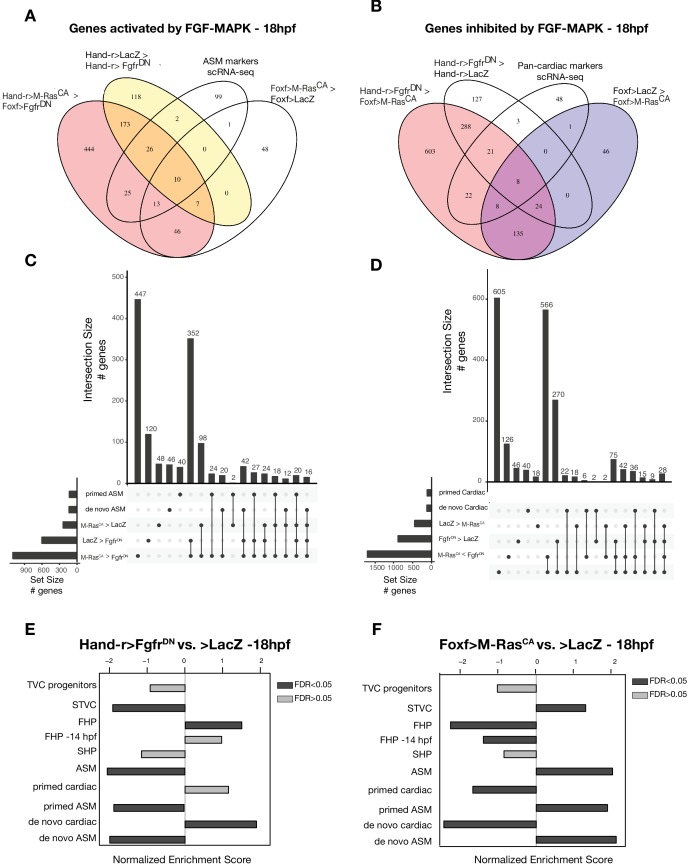
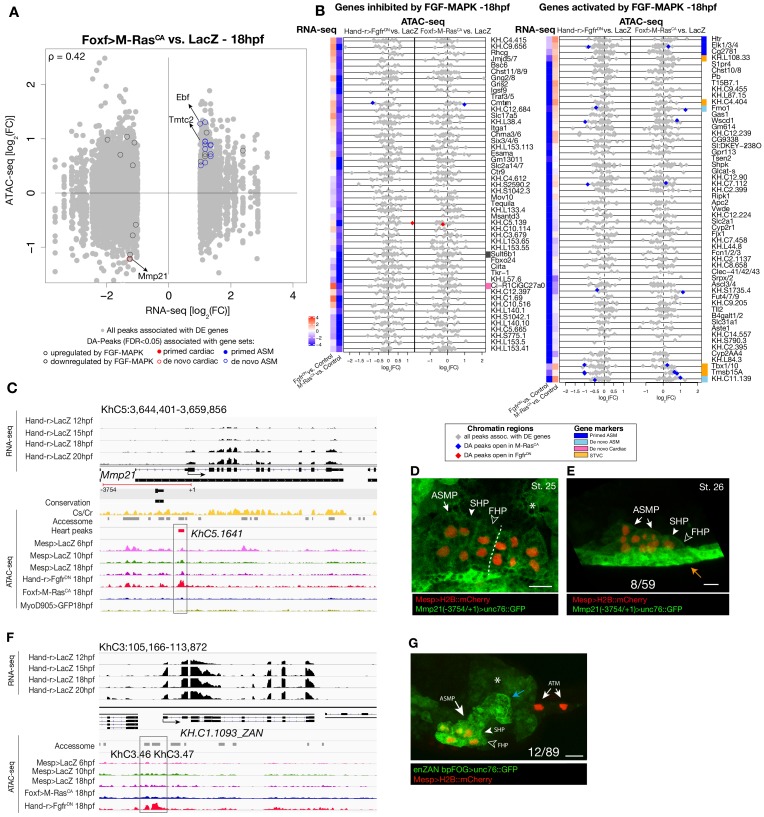
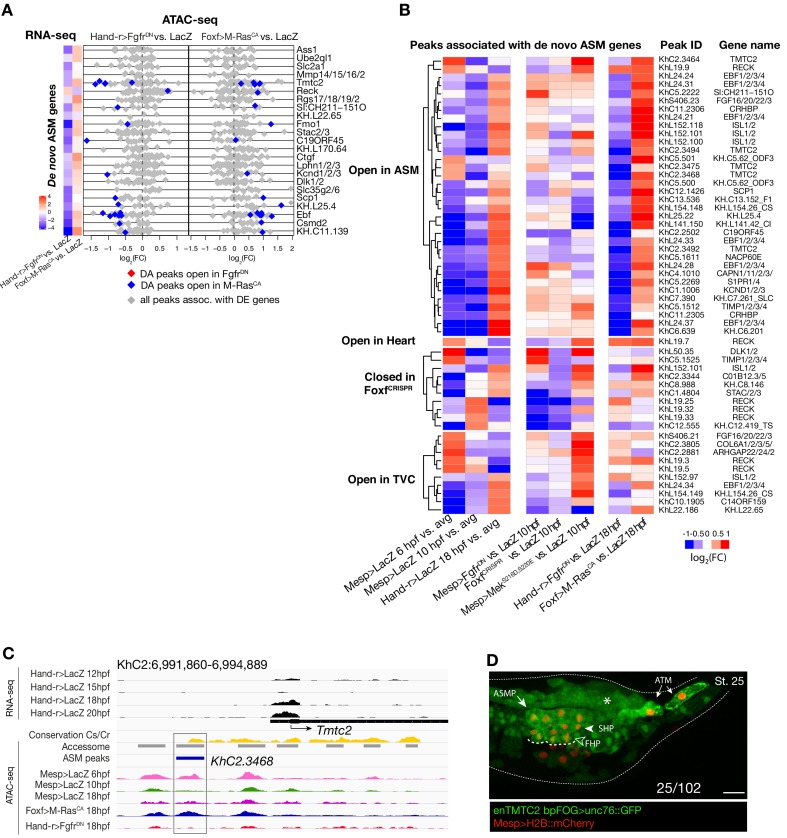
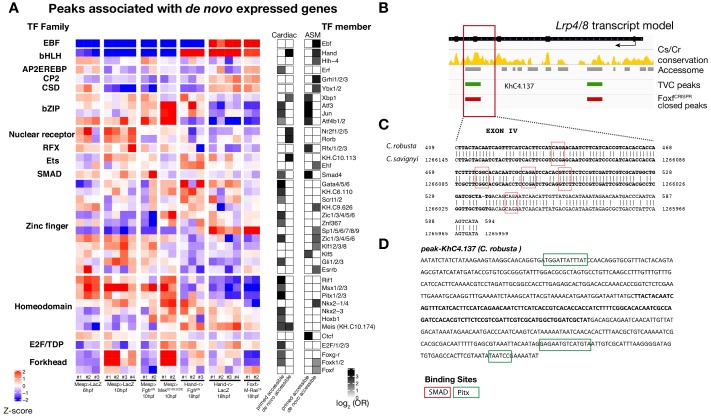
Figure 4. Cardiopharyngeal lineage-specific accessibility profiles and decoupling between enhancer accessibility and activity for de novo expressed genes.
(A) Differentially expressed (DE) genes vs. differentially accessible (DA) peaks in response to FGF-MAPK perturbation in the fate-restricted cells. ρ is the Spearman correlation of expression and accessibility for DA peaks associated with DE genes. (B) Relationship between accessibility and expression of de novo pan-cardiac genes as in Figure 2C. DE genes in either condition are shown on the left. (C) Time-dependent ATAC-seq peaks associated with de novo expressed pan-cardiac genes. The accessibility of these peaks is shown for 6, 10 and 18 hpf vs. the average accessibility in the controls (LacZ) and upon FGF-MAPK perturbations at either 10 or 18 hpf. Peaks were classified as ‘Open in ASM’ (less accessible in FgfrDN vs. M-RasCA or LacZ at 18 hpf), ‘Open in Heart’ (less accessible in M-RasCAvs. FgfrDNor LacZ at 18 hpf), ‘Closed in FoxfCRISPR’ (less accessible in FoxfCRISPR vs. ControlCRISPR), or ‘Open in TVC’ (less accessible in FgfrDN vs. MekS216D,S220E or LacZ at 10 hpf). Only regions changing accessibility between 6 and 10 hpf, or 10 and 18 hpf are shown. (D) A 6 kb region on chromosome four displaying expression profiles of RNA-seq and chromatin accessibility profiles of ATAC-seq normalized tag count. Peak ID refers to elements tested for reporter assay in vivo. The newly identified enhancer in Lrp4/8 locus is in the boxed region. (E) Enhancer-driven in vivo reporter expression (green) of tested ‘KhC4.137’ peak. TVCs marked with Mesp>H2B::mCherry (red). Numbers indicate observed/total of half-embryo scored. Zoom on cardiopharyngeal cell lineage (panel on the right). (F) Endogenous expression of Lrp4/8 visualized by in situ (green) in TyrosinaseCRISPR and upon CRISPR/Cas9-induced deletion of ATAC-seq peaks. Nuclei of B7.5 lineage cells are labelled by Mesp>nls::LacZ and revealed with an anti beta-galactosidase antibody (red). Mesp-driven hCD4::mCherry accumulates at the cell membrane as revealed by anti mCherry antibody (Blue). Experiment performed in biological replicates. Scale bar = 10 μm. (G) Fisher exact test; n is the total number of individual embryo halves scored per condition. (H) Summary model: patterns of chromatin accessibility dynamics and gene expression during early cardiopharyngeal fate specification.