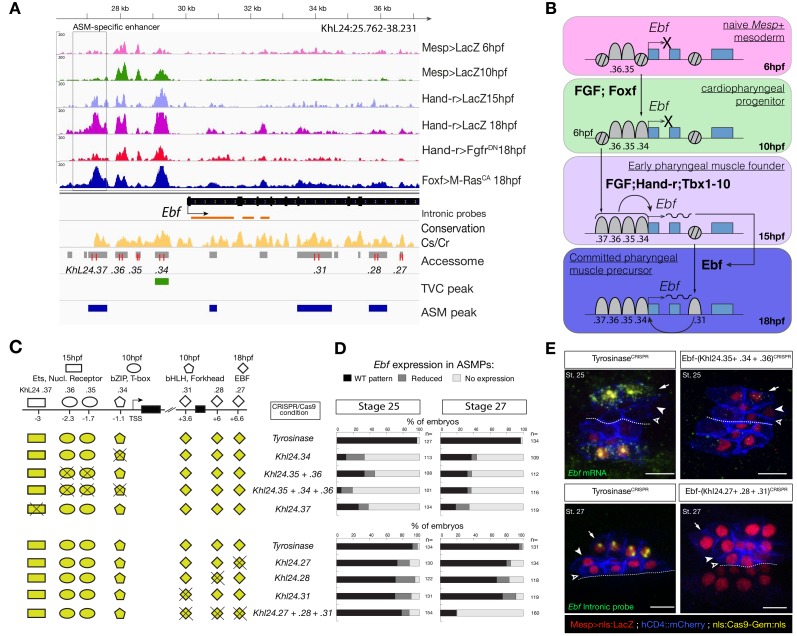
Figure 5. Combinations of cis-regulatory elements with distinct chromatin accessibility profiles are required for Ebf transcription in pharyngeal-muscle precursors.
(A) A 12 kb region of the scaffold L24 displaying expression profiles of RNA-seq and chromatin accessibility profiles of ATAC-seq (normalized tag count) in the Ebf locus. sgRNAs used to target ATAC-seq peaks are shown in red; intronic antisense riboprobes are shown in orange (B) Schematic representation showing sequential opening of cis-regulatory elements required for Ebf activation in pharyngeal muscle founder cells, and maintenance by auto-regulation in committed precursor. (C) Schematic representation of Ebf cis-regulatory elements targeted for CRISPR/Cas9-mediated deletions. Shapes represent binding sites located in the regulatory elements and differentially accessible over time. (D) Proportions of larva halves showing the indicated Ebf transcription patterns, in indicated experimental conditions; all the treatments were significant versus Tyrosinase (Fisher exact test, p < 0.001). (E) Endogenous expression of Ebf visualized by in situ (green) in TyrosinaseCRISPR and upon CRISPR/Cas9-induced deletion of ATAC-seq peaks as indicated, at stage 25 (E) and 27 (F) based on Hotta et al. (2007). For stage 25, an anti-sense riboprobe for the full length cDNA was used, whereas for stage 27 an intronic anti-sense riboprobe targeting the first three introns of Ebf transcript (orange lines) as previously used in Wang et al. (2013). Nuclei of B7.5 lineage cells are labelled by Mesp>nls::LacZ and revealed with an anti beta-galactosidase antibody (red). Mesp-driven hCD4::mCherry accumulates at the cell membrane as revealed by anti mCherry antibody (Blue). Scale bar = 10 µm.