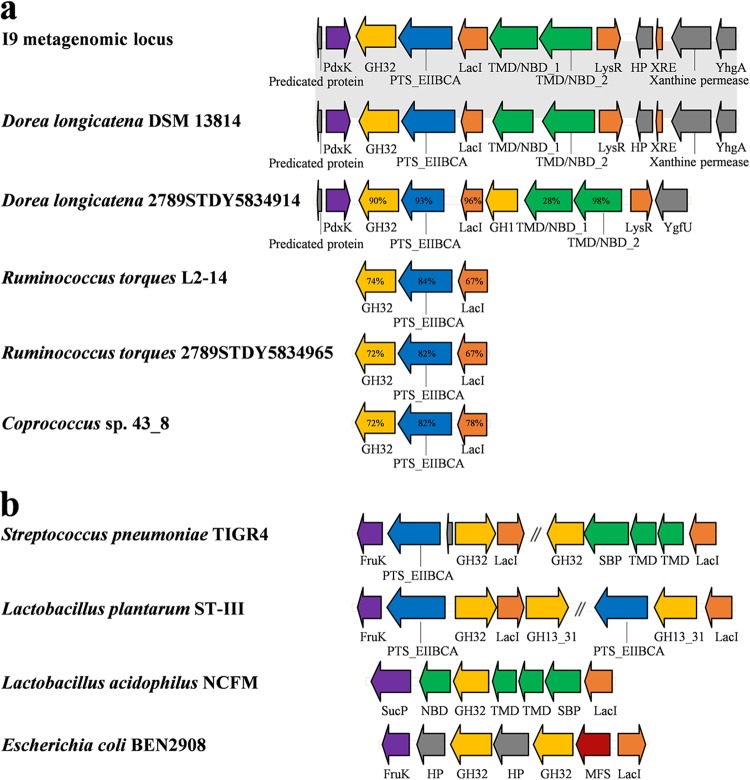
FIG 1.
Representations of FOS and sucrose utilization loci. (a) Alignment of the I9 metagenomic FOS utilization locus with similar loci from cultured bacteria. Synteny between the I9 sequence and the genomic locus of D. longicatena DSM 13814 is shown by gray bars. Percentages of identity are indicated when lower than 100%. (b) Alignment of the characterized FOS and sucrose utilization loci involving either an ABC, an MFS, or a PTS transport system, a GH32, and a related regulator. Genes predicted as being involved in carbohydrate sensing, transport, and hydrolysis are color coded: glycoside hydrolases (GH) in yellow, PTS in blue, ABC transporter components in green (NBD, nucleotide binding domain; TMD, transmembrane domain; SBP, solute binding protein), MFS in red, LacI regulator and XRE (xenobiotic response element) family transcriptional regulator in orange, and PdxK (pyridoxine kinase), FruK (fructokinase), and SucP (sucrose phosphorylases) in purple. The other genes are indicated in gray: hypothetical protein (HP), xanthine permease, YhgA (putative YhgA-like transposase), and YgfU (putative purine permease).