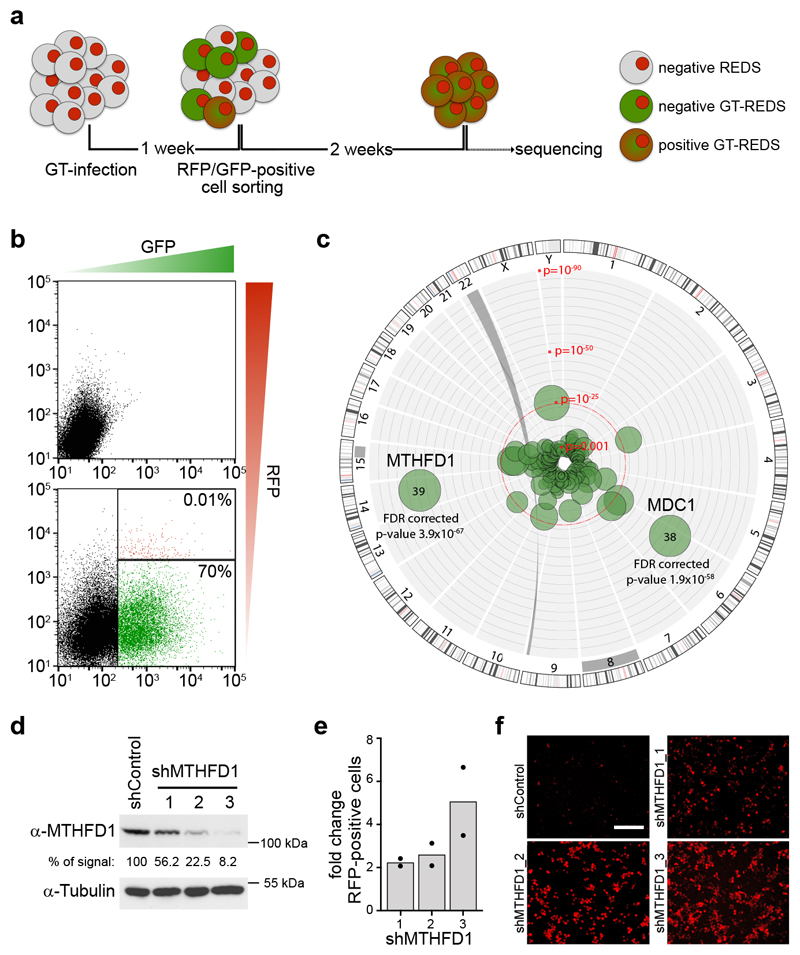
Figure 1. A genetic screen identifies MTHFD1 as a functional partner of BRD4.
a, Schematic overview of the gene-trap based genetic screen. b, Representative panels of the applied FACS-sorting strategy showing non-infected (upper panel) and gene-trap infected (lower panel) REDS1 cells; infected double-positive (GFP+/RFP+) cells (shown in red: 0.01%) were sorted. c, Circos plot illustrating the results from the gene-trap screen by genomic location (outside ring), number of independent inactivating integrations (bubble size) and significance (distance from center). P values were calculated by one-sided Fisher's exact test of insertions over an unselected control data set adjusted for false discovery rate (FDR) using Benjamini-Hochberg procedure. The screen was performed in three biologically independent experiments. d, Western blot showing MTHFD1 protein levels after downregulation with the indicated shRNAs in REDS1 cells. Numbers indicate the percentage of MTHFD1 protein remaining, tubulin was used as a loading control. The experiment was repeated three times with similar results. e, Quantification of RFP+ cells from live-cell imaging of REDS1 cells treated with MTHFD1 shRNA. Two biological replicates were done for each experimental condition. f, Representative live-cell images of MTHFD1 knock-down in REDS1 cells. Scale bar 100 μm.