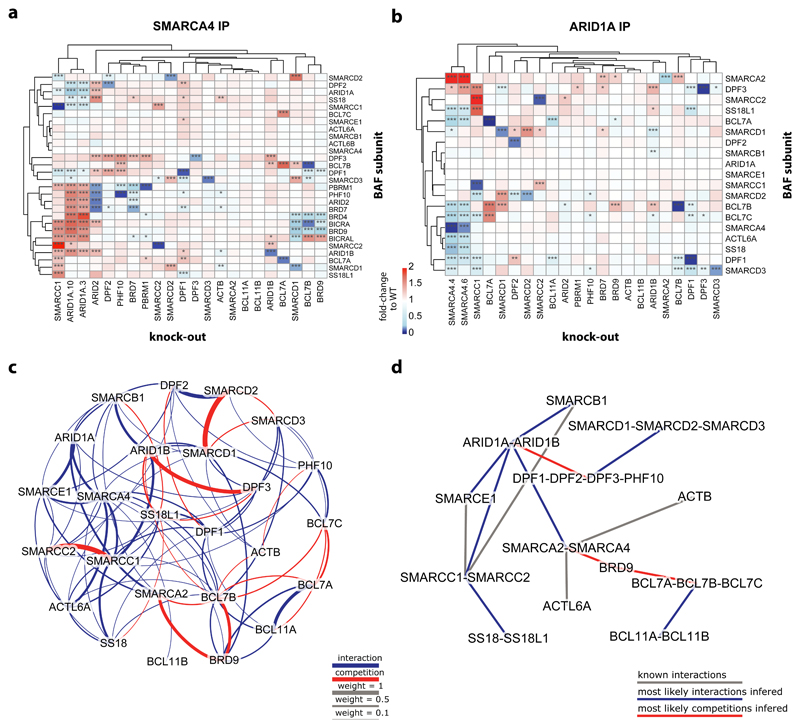
Fig. 2. BAF complex composition changes upon knock-out of single BAF-coding genes.
(a) Heatmap showing enrichment of BAF subunits in SMARCA4 immunoprecipitation in the different HAP1 knock-out cells relative to their enrichment in HAP1 WT cells. Hierarchical clustering of the knock-outs and the BAF subunits was done based on Euclidean distance. n = 2 biological independent experiments. Significant changes were calculated as described in the Supplementary Note and resulting FDR values are indicated by * (* < 1%, ** < 0.1%, *** < 0.01%). (b) Same as in a) for the results of the ARID1A IP-MS. (c) Weighted frequencies of all pairwise interactions (blue) or competitions (red) between the subunits of the BAF complexes as inferred from simulations based on the ARID1A IP-MS data. The weight of an interaction or competition is given by the normalized sum of the fitnesses of all simulated graphs in which it was observed. A cut-off of weight > 0.1095 was used, corresponding to the minimal weight at which all subunits form a single connected graph. (d) Putative organization of the BAF complexes obtained by combining interactions and competition classes with direct experimental evidence and most likely interactions inferred from the simulations based on the ARID1A IP-MS data (using a cut-off of weight > 0.42 to result in a single connected graph).