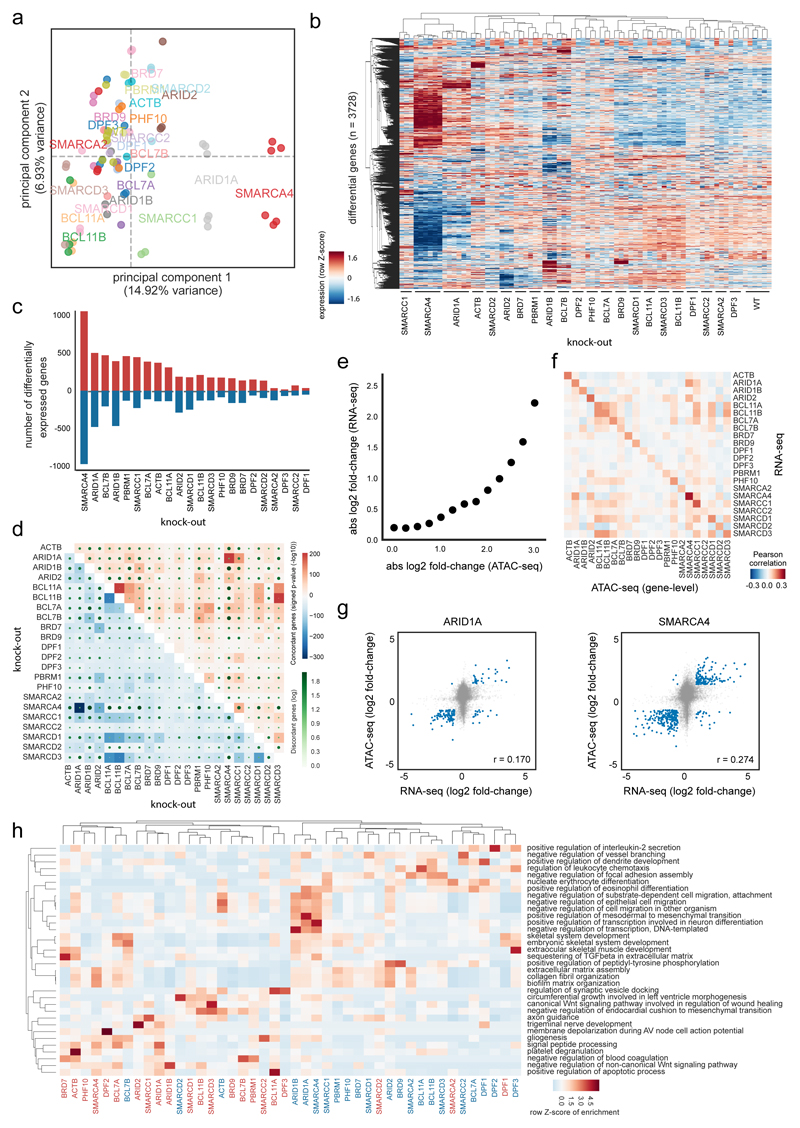
Fig. 4. Expression changes correlate with altered chromatin accessibility.
(a) Principal component analysis of RNA-seq data in HAP1 WT and KO cells (KO gene is indicated) using all expressed genes. (b) Clustering of samples based on differentially expressed genes between any KO and WT cells (KO gene is indicated). Hierarchical clustering was performed with Pearson correlation as distance measure between expression values transformed with a Z-score per row. (c) Number of up- and down-regulated genes per KO. (d) The percentage of genes that change in two KOs concordantly compared to WT are shown in blue for down-regulated and in red for up-regulated genes. The number of disconcordantly changing genes are indicated with green dots. Significance of overlap was assessed with a one-sided Fisher’s Exact Test without adjustments for multiple comparisons. (e) Relationship of expression and ATAC-seq signal aggregated per genes across all KOs. Genes were binned into groups based on ATAC-seq change. (f) Pearson correlation of log fold-changes in KO versus WT cells (KO gene indicated) between RNA-seq and ATAC-seq. (g) Scatter plot and Pearson correlation coefficient for HAP1 ARID1AKO and SMARCA4KO cells between RNA-seq and ATAC-seq log fold-changes relative to HAP1 WT cells. (h) Over-representation enrichment analysis of Gene Ontology terms for up- or down-regulated genes in different HAP1 KO cells. A row Z-score on the –log10(P value) of a one-sided Fisher’s Exact Test (Enrichr software) is displayed without adjustments for multiple comparisons. RNA-seq data of all panels are derived from n = 3 biologically independent experiments per clone.